Jean-Christophe Olivo-Marin
Paper download is intended for registered attendees only, and is
subjected to the IEEE Copyright Policy. Any other use is strongly forbidden.
Papers from this author
Extended Depth of Field Preserving Color Fidelity for Automated Digital Cytology
Alexandre Bouyssoux, Riadh Fezzani, Jean-Christophe Olivo-Marin
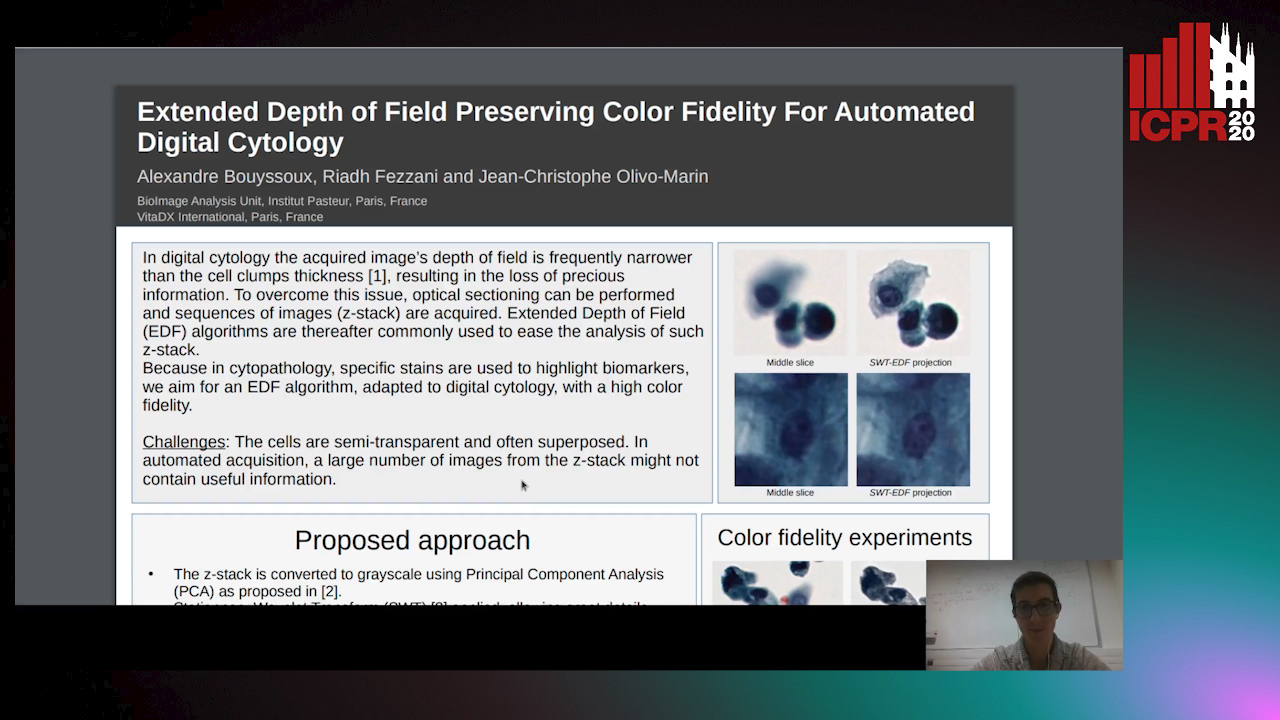
Auto-TLDR; Multi-Channel Extended Depth of Field for Digital cytology based on the stationary wavelet transform
This paper presents a multi-channel Extended Depth of Field (EDF) method for digital cytology based on the stationary wavelet transform. With a coefficient selection rule adapted to a precise color recovery, a sharp image can be reconstructed even on images with transparent overlapping cells. The precision and the color fidelity of the proposed method is analyzed. Moreover, an experiment demonstrating the necessity of volume analysis in cytology to achieve precise segmentation on cell clumps is conducted, and the importance of color fidelity in this context is asserted. The proposed method was tested on pap-stained urothelial cells and gray-scale cervical cells with important overlapping.
Learning to Segment Clustered Amoeboid Cells from Brightfield Microscopy Via Multi-Task Learning with Adaptive Weight Selection
Rituparna Sarkar, Suvadip Mukherjee, Elisabeth Labruyere, Jean-Christophe Olivo-Marin
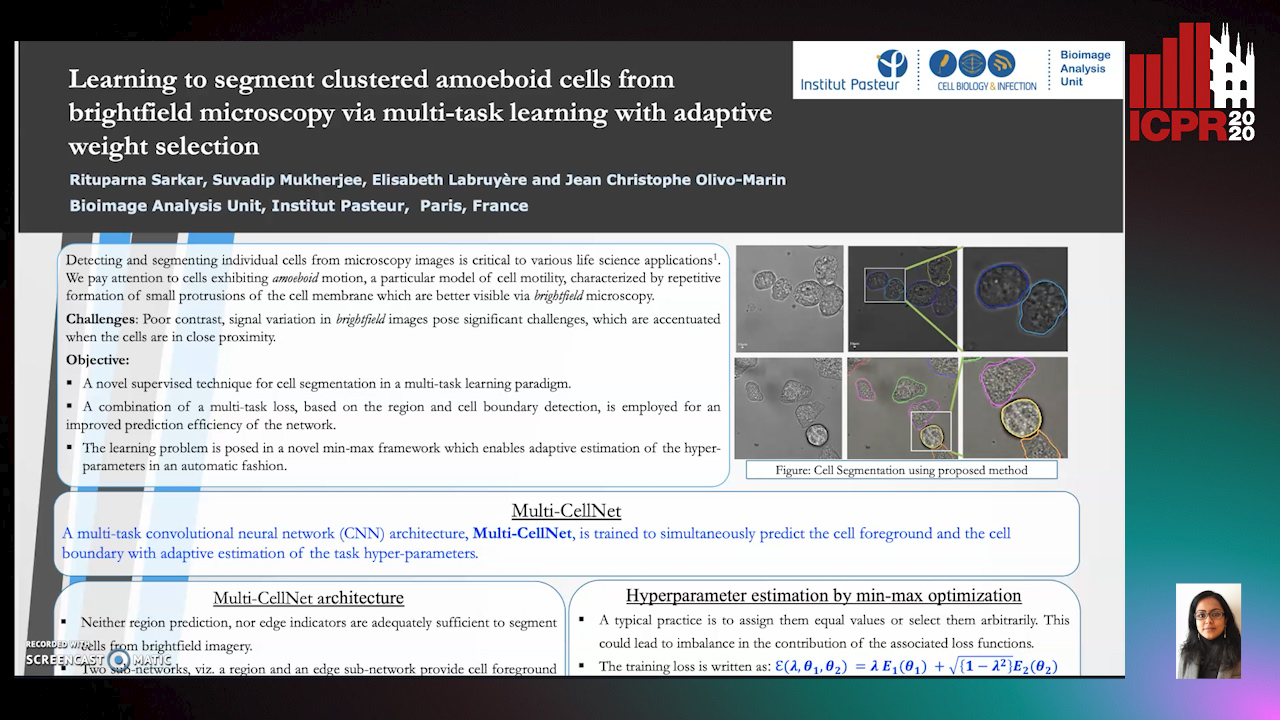
Auto-TLDR; Supervised Cell Segmentation from Microscopy Images using Multi-task Learning in a Multi-Task Learning Paradigm
Detecting and segmenting individual cells from microscopy images is critical to various life science applications. Traditional cell segmentation tools are often ill-suited for applications in brightfield microscopy due to poor contrast and intensity heterogeneity, and only a small subset are applicable to segment cells in a cluster. In this regard, we introduce a novel supervised technique for cell segmentation in a multi-task learning paradigm. A combination of a multi-task loss, based on the region and cell boundary detection, is employed for an improved prediction efficiency of the network. The learning problem is posed in a novel min-max framework which enables adaptive estimation of the hyper-parameters in an automatic fashion. The region and cell boundary predictions are combined via morphological operations and active contour model to segment individual cells. The proposed methodology is particularly suited to segment touching cells from brightfield microscopy images without manual interventions. Quantitatively, we observe an overall Dice score of 0.93 on the validation set, which is an improvement of over 15.9% on a recent unsupervised method, and outperforms the popular supervised U-net algorithm by at least 5.8% on average.