A Deep Learning Approach for the Segmentation of Myocardial Diseases
Khawala Brahim,
Abdull Qayyum,
Alain Lalande,
Arnaud Boucher,
Anis Sakly,
Fabrice Meriaudeau
Auto-TLDR; Segmentation of Myocardium Infarction Using Late GADEMRI and SegU-Net
Similar papers
FOANet: A Focus of Attention Network with Application to Myocardium Segmentation
Zhou Zhao, Elodie Puybareau, Nicolas Boutry, Thierry Geraud

Auto-TLDR; FOANet: A Hybrid Loss Function for Myocardium Segmentation of Cardiac Magnetic Resonance Images
Abstract Slides Poster Similar
Automatic Semantic Segmentation of Structural Elements related to the Spinal Cord in the Lumbar Region by Using Convolutional Neural Networks
Jhon Jairo Sáenz Gamboa, Maria De La Iglesia-Vaya, Jon Ander Gómez

Auto-TLDR; Semantic Segmentation of Lumbar Spine Using Convolutional Neural Networks
Abstract Slides Poster Similar
A Benchmark Dataset for Segmenting Liver, Vasculature and Lesions from Large-Scale Computed Tomography Data
Bo Wang, Zhengqing Xu, Wei Xu, Qingsen Yan, Liang Zhang, Zheng You

Auto-TLDR; The Biggest Treatment-Oriented Liver Cancer Dataset for Segmentation
Abstract Slides Poster Similar
Deep Recurrent-Convolutional Model for AutomatedSegmentation of Craniomaxillofacial CT Scans
Francesca Murabito, Simone Palazzo, Federica Salanitri Proietto, Francesco Rundo, Ulas Bagci, Daniela Giordano, Rosalia Leonardi, Concetto Spampinato
Auto-TLDR; Automated Segmentation of Anatomical Structures in Craniomaxillofacial CT Scans using Fully Convolutional Deep Networks
Abstract Slides Poster Similar
Planar 3D Transfer Learning for End to End Unimodal MRI Unbalanced Data Segmentation
Martin Kolarik, Radim Burget, Carlos M. Travieso-Gonzalez, Jan Kocica

Auto-TLDR; Planar 3D Res-U-Net Network for Unbalanced 3D Image Segmentation using Fluid Attenuation Inversion Recover
A Lumen Segmentation Method in Ureteroscopy Images Based on a Deep Residual U-Net Architecture
Jorge Lazo, Marzullo Aldo, Sara Moccia, Michele Catellani, Benoit Rosa, Elena De Momi, Michel De Mathelin, Francesco Calimeri
Auto-TLDR; A Deep Neural Network for Ureteroscopy with Residual Units
Abstract Slides Poster Similar
Do Not Treat Boundaries and Regions Differently: An Example on Heart Left Atrial Segmentation
Zhou Zhao, Elodie Puybareau, Nicolas Boutry, Thierry Geraud
Auto-TLDR; Attention Full Convolutional Network for Atrial Segmentation using ResNet-101 Architecture
BG-Net: Boundary-Guided Network for Lung Segmentation on Clinical CT Images
Rui Xu, Yi Wang, Tiantian Liu, Xinchen Ye, Lin Lin, Yen-Wei Chen, Shoji Kido, Noriyuki Tomiyama
Auto-TLDR; Boundary-Guided Network for Lung Segmentation on CT Images
Abstract Slides Poster Similar
One-Stage Multi-Task Detector for 3D Cardiac MR Imaging
Weizeng Lu, Xi Jia, Wei Chen, Nicolò Savioli, Antonio De Marvao, Linlin Shen, Declan O'Regan, Jinming Duan

Auto-TLDR; Multi-task Learning for Real-Time, simultaneous landmark location and bounding box detection in 3D space
Abstract Slides Poster Similar
End-To-End Multi-Task Learning for Lung Nodule Segmentation and Diagnosis
Wei Chen, Qiuli Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; A novel multi-task framework for lung nodule diagnosis based on deep learning and medical features
Segmentation of Intracranial Aneurysm Remnant in MRA Using Dual-Attention Atrous Net
Subhashis Banerjee, Ashis Kumar Dhara, Johan Wikström, Robin Strand
Auto-TLDR; Dual-Attention Atrous Net for Segmentation of Intracranial Aneurysm Remnant from MRA Images
Abstract Slides Poster Similar
MTGAN: Mask and Texture-Driven Generative Adversarial Network for Lung Nodule Segmentation
Wei Chen, Qiuli Wang, Kun Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; Mask and Texture-driven Generative Adversarial Network for Lung Nodule Segmentation
Abstract Slides Poster Similar
CAggNet: Crossing Aggregation Network for Medical Image Segmentation
Auto-TLDR; Crossing Aggregation Network for Medical Image Segmentation
Abstract Slides Poster Similar
BiLuNet: A Multi-Path Network for Semantic Segmentation on X-Ray Images
Van Luan Tran, Huei-Yung Lin, Rachel Liu, Chun-Han Tseng, Chun-Han Tseng
Auto-TLDR; BiLuNet: Multi-path Convolutional Neural Network for Semantic Segmentation of Lumbar vertebrae, sacrum,
A Multi-Task Contextual Atrous Residual Network for Brain Tumor Detection & Segmentation
Ngan Le, Kashu Yamazaki, Quach Kha Gia, Thanh-Dat Truong, Marios Savvides

Auto-TLDR; Contextual Brain Tumor Segmentation Using 3D atrous Residual Networks and Cascaded Structures
Unsupervised Detection of Pulmonary Opacities for Computer-Aided Diagnosis of COVID-19 on CT Images
Rui Xu, Xiao Cao, Yufeng Wang, Yen-Wei Chen, Xinchen Ye, Lin Lin, Wenchao Zhu, Chao Chen, Fangyi Xu, Yong Zhou, Hongjie Hu, Shoji Kido, Noriyuki Tomiyama
Auto-TLDR; A computer-aided diagnosis of COVID-19 from CT images using unsupervised pulmonary opacity detection
Abstract Slides Poster Similar
BCAU-Net: A Novel Architecture with Binary Channel Attention Module for MRI Brain Segmentation
Yongpei Zhu, Zicong Zhou, Guojun Liao, Kehong Yuan

Auto-TLDR; BCAU-Net: Binary Channel Attention U-Net for MRI brain segmentation
Abstract Slides Poster Similar
Transfer Learning through Weighted Loss Function and Group Normalization for Vessel Segmentation from Retinal Images
Abdullah Sarhan, Jon Rokne, Reda Alhajj, Andrew Crichton

Auto-TLDR; Deep Learning for Segmentation of Blood Vessels in Retinal Images
Abstract Slides Poster Similar
Deep Learning-Based Type Identification of Volumetric MRI Sequences
Jean Pablo De Mello, Thiago Paixão, Rodrigo Berriel, Mauricio Reyes, Alberto F. De Souza, Claudine Badue, Thiago Oliveira-Santos

Auto-TLDR; Deep Learning for Brain MRI Sequences Identification Using Convolutional Neural Network
Abstract Slides Poster Similar
DE-Net: Dilated Encoder Network for Automated Tongue Segmentation
Hui Tang, Bin Wang, Jun Zhou, Yongsheng Gao

Auto-TLDR; Automated Tongue Image Segmentation using De-Net
Abstract Slides Poster Similar
EM-Net: Deep Learning for Electron Microscopy Image Segmentation
Afshin Khadangi, Thomas Boudier, Vijay Rajagopal

Auto-TLDR; EM-net: Deep Convolutional Neural Network for Electron Microscopy Image Segmentation
3D Medical Multi-Modal Segmentation Network Guided by Multi-Source Correlation Constraint
Tongxue Zhou, Stéphane Canu, Pierre Vera, Su Ruan
Auto-TLDR; Multi-modality Segmentation with Correlation Constrained Network
Abstract Slides Poster Similar
NephCNN: A Deep-Learning Framework for Vessel Segmentation in Nephrectomy Laparoscopic Videos
Alessandro Casella, Sara Moccia, Chiara Carlini, Emanuele Frontoni, Elena De Momi, Leonardo Mattos

Auto-TLDR; Adversarial Fully Convolutional Neural Networks for kidney vessel segmentation from nephrectomy laparoscopic videos
Abstract Slides Poster Similar
DA-RefineNet: Dual-Inputs Attention RefineNet for Whole Slide Image Segmentation
Ziqiang Li, Rentuo Tao, Qianrun Wu, Bin Li

Auto-TLDR; DA-RefineNet: A dual-inputs attention network for whole slide image segmentation
Abstract Slides Poster Similar
A Deep Learning-Based Method for Predicting Volumes of Nasopharyngeal Carcinoma for Adaptive Radiation Therapy Treatment
Bilel Daoud, Ken'Ichi Morooka, Shoko Miyauchi, Ryo Kurazume, Wafa Mnejja, Leila Farhat, Jamel Daoud

Auto-TLDR; TEP-Net: Tumor Evolution Prediction of Nasopharyngeal Carcinoma and Organ-at-risks Using CT Images
Abstract Slides Poster Similar
Dual Encoder Fusion U-Net (DEFU-Net) for Cross-manufacturer Chest X-Ray Segmentation
Zhang Lipei, Aozhi Liu, Jing Xiao
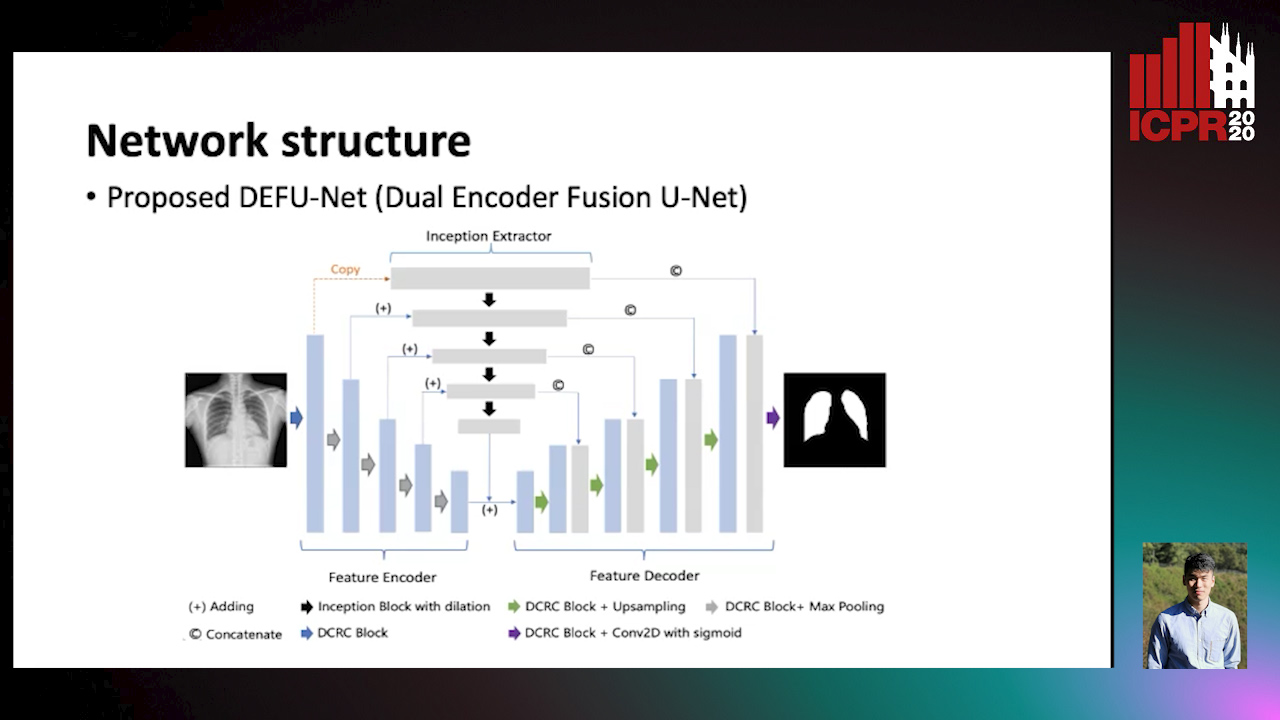
Auto-TLDR; Inception Convolutional Neural Network with Dilation for Chest X-Ray Segmentation
Triplet-Path Dilated Network for Detection and Segmentation of General Pathological Images
Jiaqi Luo, Zhicheng Zhao, Fei Su, Limei Guo

Auto-TLDR; Triplet-path Network for One-Stage Object Detection and Segmentation in Pathological Images
Leveraging Unlabeled Data for Glioma Molecular Subtype and Survival Prediction
Nicholas Nuechterlein, Beibin Li, Mehmet Saygin Seyfioglu, Sachin Mehta, Patrick Cimino, Linda Shapiro

Auto-TLDR; Multimodal Brain Tumor Segmentation Using Unlabeled MR Data and Genomic Data for Cancer Prediction
Abstract Slides Poster Similar
Learn to Segment Retinal Lesions and Beyond
Qijie Wei, Xirong Li, Weihong Yu, Xiao Zhang, Yongpeng Zhang, Bojie Hu, Bin Mo, Di Gong, Ning Chen, Dayong Ding, Youxin Chen

Auto-TLDR; Multi-task Lesion Segmentation and Disease Classification for Diabetic Retinopathy Grading
Segmentation of Axillary and Supraclavicular Tumoral Lymph Nodes in PET/CT: A Hybrid CNN/Component-Tree Approach
Diana Lucia Farfan Cabrera, Nicolas Gogin, David Morland, Benoît Naegel, Dimitri Papathanassiou, Nicolas Passat

Auto-TLDR; Coupling Convolutional Neural Networks and Component-Trees for Lymph node Segmentation from PET/CT Images
OCT Image Segmentation Using NeuralArchitecture Search and SRGAN
Saba Heidari, Omid Dehzangi, Nasser M. Nasarabadi, Ali Rezai
Auto-TLDR; Automatic Segmentation of Retinal Layers in Optical Coherence Tomography using Neural Architecture Search
Segmenting Kidney on Multiple Phase CT Images Using ULBNet
Yanling Chi, Yuyu Xu, Gang Feng, Jiawei Mao, Sihua Wu, Guibin Xu, Weimin Huang
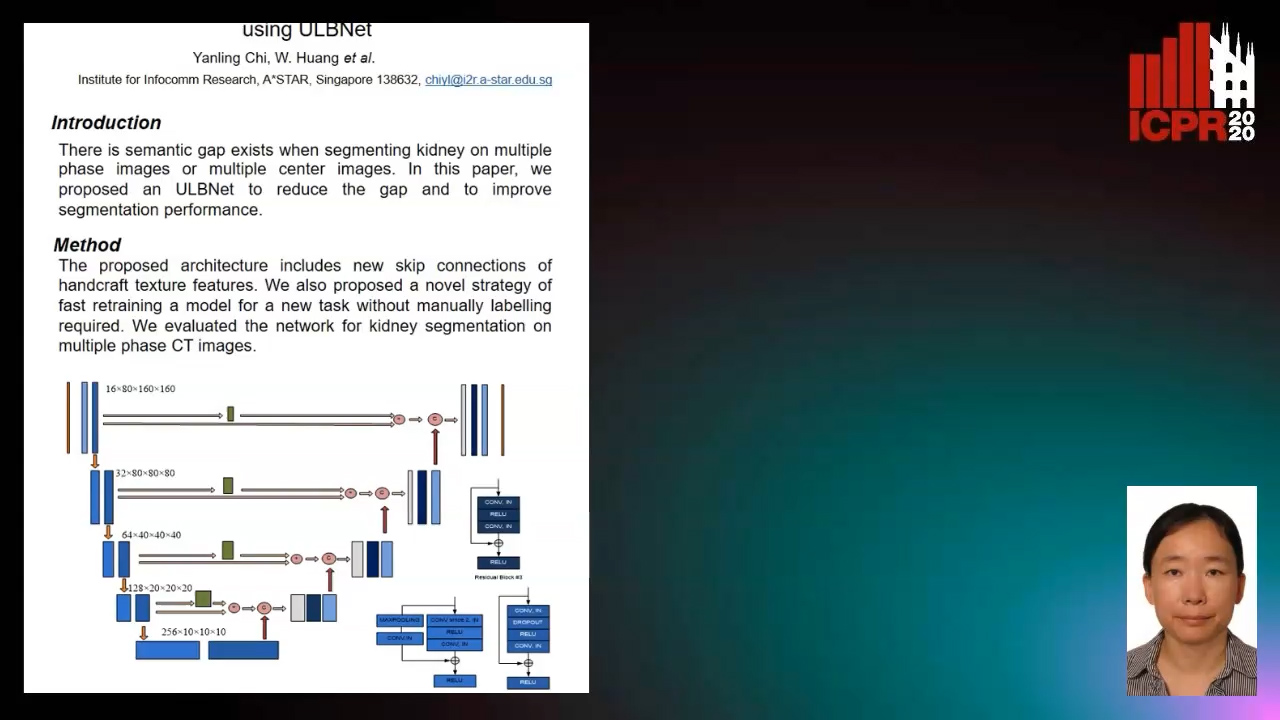
Auto-TLDR; A ULBNet network for kidney segmentation on multiple phase CT images
Offset Curves Loss for Imbalanced Problem in Medical Segmentation
Ngan Le, Duc Toan Bui, Khoa Luu, Marios Savvides

Auto-TLDR; Offset Curves Loss for Medical Image Segmentation
Learning to Segment Clustered Amoeboid Cells from Brightfield Microscopy Via Multi-Task Learning with Adaptive Weight Selection
Rituparna Sarkar, Suvadip Mukherjee, Elisabeth Labruyere, Jean-Christophe Olivo-Marin
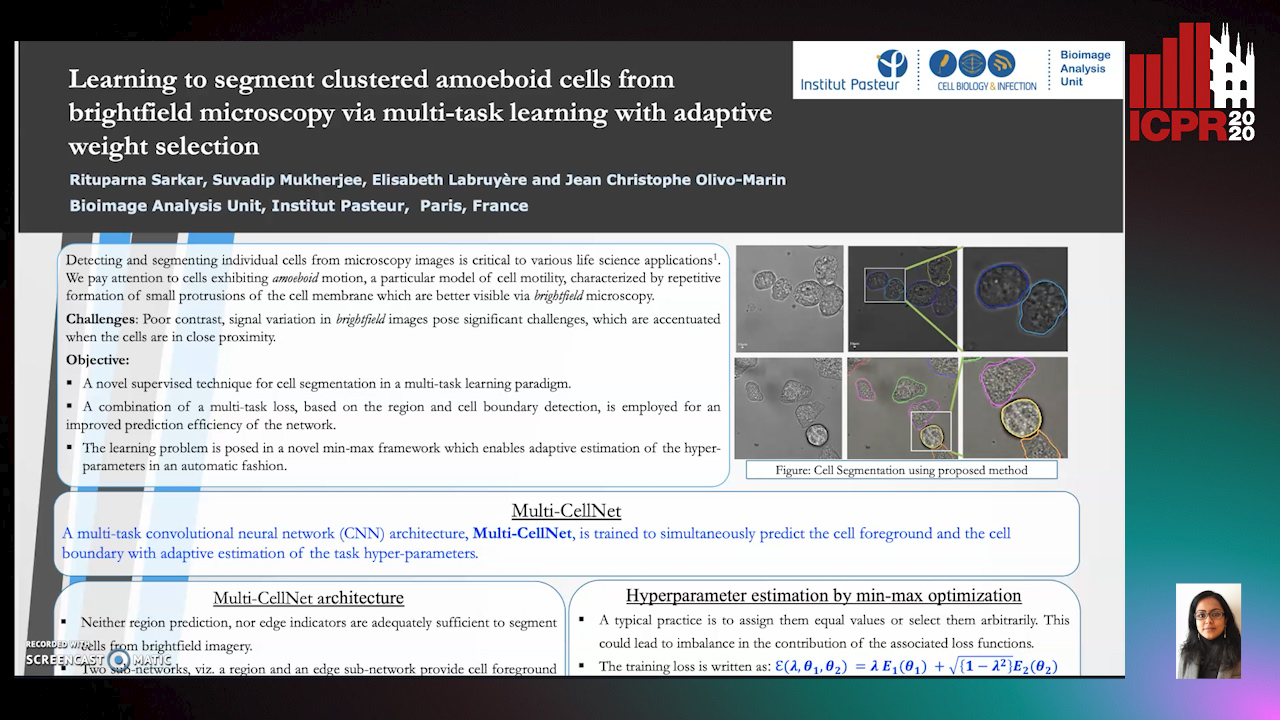
Auto-TLDR; Supervised Cell Segmentation from Microscopy Images using Multi-task Learning in a Multi-Task Learning Paradigm
Accurate Cell Segmentation in Digital Pathology Images Via Attention Enforced Networks
Zeyi Yao, Kaiqi Li, Guanhong Zhang, Yiwen Luo, Xiaoguang Zhou, Muyi Sun

Auto-TLDR; AENet: Attention Enforced Network for Automatic Cell Segmentation
Abstract Slides Poster Similar
CT-UNet: An Improved Neural Network Based on U-Net for Building Segmentation in Remote Sensing Images
Huanran Ye, Sheng Liu, Kun Jin, Haohao Cheng
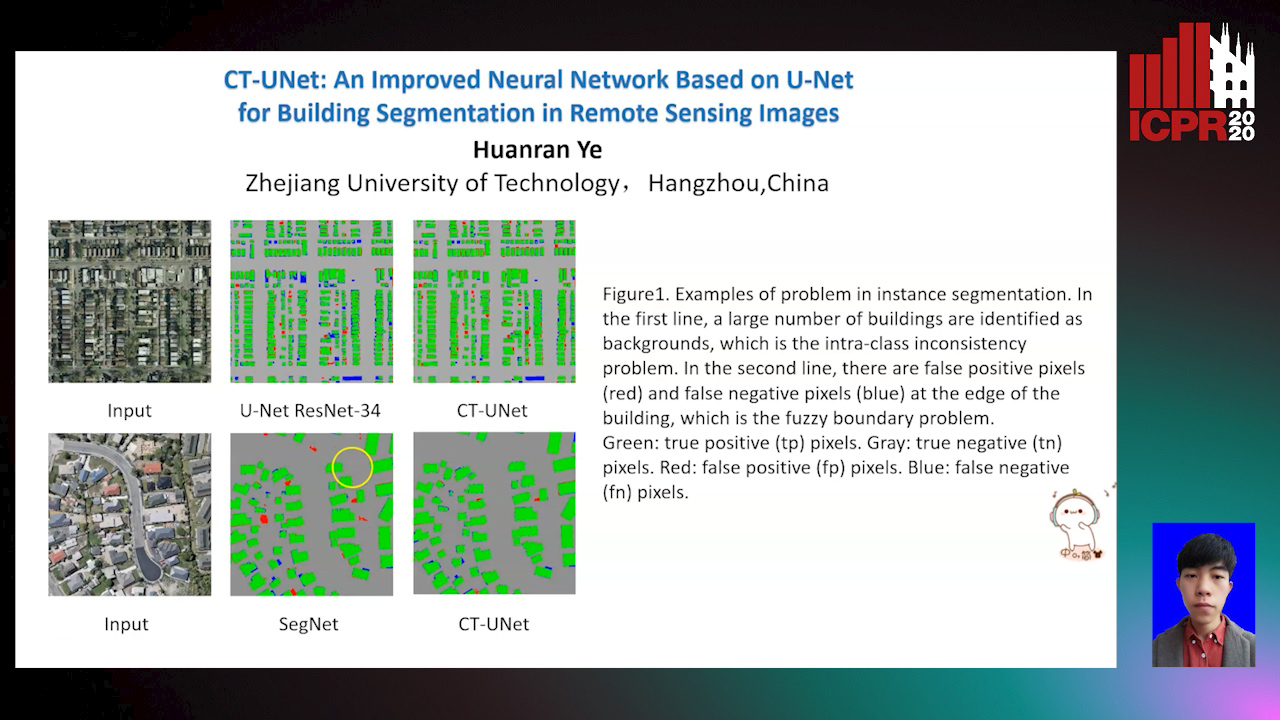
Auto-TLDR; Context-Transfer-UNet: A UNet-based Network for Building Segmentation in Remote Sensing Images
Abstract Slides Poster Similar
Fine-Tuning Convolutional Neural Networks: A Comprehensive Guide and Benchmark Analysis for Glaucoma Screening
Amed Mvoulana, Rostom Kachouri, Mohamed Akil
Auto-TLDR; Fine-tuning Convolutional Neural Networks for Glaucoma Screening
Abstract Slides Poster Similar
A Transformer-Based Network for Anisotropic 3D Medical Image Segmentation
Guo Danfeng, Demetri Terzopoulos
Auto-TLDR; A transformer-based model to tackle the anisotropy problem in 3D medical image analysis
Abstract Slides Poster Similar
Vesselness Filters: A Survey with Benchmarks Applied to Liver Imaging
Jonas Lamy, Odyssée Merveille, Bertrand Kerautret, Nicolas Passat, Antoine Vacavant

Auto-TLDR; Comparison of Vessel Enhancement Filters for Liver Vascular Network Segmentation
Abstract Slides Poster Similar
A Systematic Investigation on Deep Architectures for Automatic Skin Lesions Classification
Pierluigi Carcagni, Marco Leo, Andrea Cuna, Giuseppe Celeste, Cosimo Distante

Auto-TLDR; RegNet: Deep Investigation of Convolutional Neural Networks for Automatic Classification of Skin Lesions
Abstract Slides Poster Similar
Prediction of Obstructive Coronary Artery Disease from Myocardial Perfusion Scintigraphy using Deep Neural Networks
Ida Arvidsson, Niels Christian Overgaard, Miguel Ochoa Figueroa, Jeronimo Rose, Anette Davidsson, Kalle Åström, Anders Heyden

Auto-TLDR; A Deep Learning Algorithm for Multi-label Classification of Myocardial Perfusion Scintigraphy for Stable Ischemic Heart Disease
Abstract Slides Poster Similar
Neural Machine Registration for Motion Correction in Breast DCE-MRI
Federica Aprea, Stefano Marrone, Carlo Sansone
Auto-TLDR; A Neural Registration Network for Dynamic Contrast Enhanced-Magnetic Resonance Imaging
Abstract Slides Poster Similar
Multi-focus Image Fusion for Confocal Microscopy Using U-Net Regression Map
Md Maruf Hossain Shuvo, Yasmin M. Kassim, Filiz Bunyak, Olga V. Glinskii, Leike Xie, Vladislav V Glinsky, Virginia H. Huxley, Kannappan Palaniappan

Auto-TLDR; Independent Single Channel U-Net Fusion for Multi-focus Microscopy Images
Abstract Slides Poster Similar
Dual Stream Network with Selective Optimization for Skin Disease Recognition in Consumer Grade Images
Krishnam Gupta, Jaiprasad Rampure, Monu Krishnan, Ajit Narayanan, Nikhil Narayan
Auto-TLDR; A Deep Network Architecture for Skin Disease Localisation and Classification on Consumer Grade Images
Abstract Slides Poster Similar
Fused 3-Stage Image Segmentation for Pleural Effusion Cell Clusters
Sike Ma, Meng Zhao, Hao Wang, Fan Shi, Xuguo Sun, Shengyong Chen, Hong-Ning Dai

Auto-TLDR; Coarse Segmentation of Stained and Stained Unstained Cell Clusters in pleural effusion using 3-stage segmentation method
Abstract Slides Poster Similar
Semantic Segmentation of Breast Ultrasound Image with Pyramid Fuzzy Uncertainty Reduction and Direction Connectedness Feature
Kuan Huang, Yingtao Zhang, Heng-Da Cheng, Ping Xing, Boyu Zhang

Auto-TLDR; Uncertainty-Based Deep Learning for Breast Ultrasound Image Segmentation
Abstract Slides Poster Similar
Classify Breast Histopathology Images with Ductal Instance-Oriented Pipeline
Beibin Li, Ezgi Mercan, Sachin Mehta, Stevan Knezevich, Corey Arnold, Donald Weaver, Joann Elmore, Linda Shapiro

Auto-TLDR; DIOP: Ductal Instance-Oriented Pipeline for Diagnostic Classification
Abstract Slides Poster Similar
SA-UNet: Spatial Attention U-Net for Retinal Vessel Segmentation
Changlu Guo, Marton Szemenyei, Yugen Yi, Wenle Wang, Buer Chen, Changqi Fan

Auto-TLDR; Spatial Attention U-Net for Segmentation of Retinal Blood Vessels
Abstract Slides Poster Similar