Vesselness Filters: A Survey with Benchmarks Applied to Liver Imaging
Jonas Lamy,
Odyssée Merveille,
Bertrand Kerautret,
Nicolas Passat,
Antoine Vacavant

Auto-TLDR; Comparison of Vessel Enhancement Filters for Liver Vascular Network Segmentation
Similar papers
A Benchmark Dataset for Segmenting Liver, Vasculature and Lesions from Large-Scale Computed Tomography Data
Bo Wang, Zhengqing Xu, Wei Xu, Qingsen Yan, Liang Zhang, Zheng You

Auto-TLDR; The Biggest Treatment-Oriented Liver Cancer Dataset for Segmentation
Abstract Slides Poster Similar
Segmentation of Axillary and Supraclavicular Tumoral Lymph Nodes in PET/CT: A Hybrid CNN/Component-Tree Approach
Diana Lucia Farfan Cabrera, Nicolas Gogin, David Morland, Benoît Naegel, Dimitri Papathanassiou, Nicolas Passat

Auto-TLDR; Coupling Convolutional Neural Networks and Component-Trees for Lymph node Segmentation from PET/CT Images
Segmentation of Intracranial Aneurysm Remnant in MRA Using Dual-Attention Atrous Net
Subhashis Banerjee, Ashis Kumar Dhara, Johan Wikström, Robin Strand
Auto-TLDR; Dual-Attention Atrous Net for Segmentation of Intracranial Aneurysm Remnant from MRA Images
Abstract Slides Poster Similar
Transfer Learning through Weighted Loss Function and Group Normalization for Vessel Segmentation from Retinal Images
Abdullah Sarhan, Jon Rokne, Reda Alhajj, Andrew Crichton

Auto-TLDR; Deep Learning for Segmentation of Blood Vessels in Retinal Images
Abstract Slides Poster Similar
A Novel Computer-Aided Diagnostic System for Early Assessment of Hepatocellular Carcinoma
Ahmed Alksas, Mohamed Shehata, Gehad Saleh, Ahmed Shaffie, Ahmed Soliman, Mohammed Ghazal, Hadil Abukhalifeh, Abdel Razek Ahmed, Ayman El-Baz

Auto-TLDR; Classification of Liver Tumor Lesions from CE-MRI Using Structured Structural Features and Functional Features
Abstract Slides Poster Similar
Automatic Semantic Segmentation of Structural Elements related to the Spinal Cord in the Lumbar Region by Using Convolutional Neural Networks
Jhon Jairo Sáenz Gamboa, Maria De La Iglesia-Vaya, Jon Ander Gómez

Auto-TLDR; Semantic Segmentation of Lumbar Spine Using Convolutional Neural Networks
Abstract Slides Poster Similar
Multi-focus Image Fusion for Confocal Microscopy Using U-Net Regression Map
Md Maruf Hossain Shuvo, Yasmin M. Kassim, Filiz Bunyak, Olga V. Glinskii, Leike Xie, Vladislav V Glinsky, Virginia H. Huxley, Kannappan Palaniappan

Auto-TLDR; Independent Single Channel U-Net Fusion for Multi-focus Microscopy Images
Abstract Slides Poster Similar
PCANet: Pyramid Context-Aware Network for Retinal Vessel Segmentation
Yi Zhang, Yixuan Chen, Kai Zhang

Auto-TLDR; PCANet: Adaptive Context-Aware Network for Automated Retinal Vessel Segmentation
Abstract Slides Poster Similar
A Deep Learning Approach for the Segmentation of Myocardial Diseases
Khawala Brahim, Abdull Qayyum, Alain Lalande, Arnaud Boucher, Anis Sakly, Fabrice Meriaudeau
Auto-TLDR; Segmentation of Myocardium Infarction Using Late GADEMRI and SegU-Net
Abstract Slides Poster Similar
NephCNN: A Deep-Learning Framework for Vessel Segmentation in Nephrectomy Laparoscopic Videos
Alessandro Casella, Sara Moccia, Chiara Carlini, Emanuele Frontoni, Elena De Momi, Leonardo Mattos

Auto-TLDR; Adversarial Fully Convolutional Neural Networks for kidney vessel segmentation from nephrectomy laparoscopic videos
Abstract Slides Poster Similar
FOANet: A Focus of Attention Network with Application to Myocardium Segmentation
Zhou Zhao, Elodie Puybareau, Nicolas Boutry, Thierry Geraud

Auto-TLDR; FOANet: A Hybrid Loss Function for Myocardium Segmentation of Cardiac Magnetic Resonance Images
Abstract Slides Poster Similar
3D Medical Multi-Modal Segmentation Network Guided by Multi-Source Correlation Constraint
Tongxue Zhou, Stéphane Canu, Pierre Vera, Su Ruan
Auto-TLDR; Multi-modality Segmentation with Correlation Constrained Network
Abstract Slides Poster Similar
DARN: Deep Attentive Refinement Network for Liver Tumor Segmentation from 3D CT Volume
Yao Zhang, Jiang Tian, Cheng Zhong, Yang Zhang, Zhongchao Shi, Zhiqiang He

Auto-TLDR; Deep Attentive Refinement Network for Liver Tumor Segmentation from 3D Computed Tomography Using Multi-Level Features
Abstract Slides Poster Similar
BCAU-Net: A Novel Architecture with Binary Channel Attention Module for MRI Brain Segmentation
Yongpei Zhu, Zicong Zhou, Guojun Liao, Kehong Yuan

Auto-TLDR; BCAU-Net: Binary Channel Attention U-Net for MRI brain segmentation
Abstract Slides Poster Similar
A Comparison of Neural Network Approaches for Melanoma Classification
Maria Frasca, Michele Nappi, Michele Risi, Genoveffa Tortora, Alessia Auriemma Citarella
Auto-TLDR; Classification of Melanoma Using Deep Neural Network Methodologies
Abstract Slides Poster Similar
SAGE: Sequential Attribute Generator for Analyzing Glioblastomas Using Limited Dataset
Padmaja Jonnalagedda, Brent Weinberg, Jason Allen, Taejin Min, Shiv Bhanu, Bir Bhanu

Auto-TLDR; SAGE: Generative Adversarial Networks for Imaging Biomarker Detection and Prediction
Abstract Slides Poster Similar
A Multi-Task Contextual Atrous Residual Network for Brain Tumor Detection & Segmentation
Ngan Le, Kashu Yamazaki, Quach Kha Gia, Thanh-Dat Truong, Marios Savvides

Auto-TLDR; Contextual Brain Tumor Segmentation Using 3D atrous Residual Networks and Cascaded Structures
Planar 3D Transfer Learning for End to End Unimodal MRI Unbalanced Data Segmentation
Martin Kolarik, Radim Burget, Carlos M. Travieso-Gonzalez, Jan Kocica

Auto-TLDR; Planar 3D Res-U-Net Network for Unbalanced 3D Image Segmentation using Fluid Attenuation Inversion Recover
Neural Machine Registration for Motion Correction in Breast DCE-MRI
Federica Aprea, Stefano Marrone, Carlo Sansone
Auto-TLDR; A Neural Registration Network for Dynamic Contrast Enhanced-Magnetic Resonance Imaging
Abstract Slides Poster Similar
MTGAN: Mask and Texture-Driven Generative Adversarial Network for Lung Nodule Segmentation
Wei Chen, Qiuli Wang, Kun Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; Mask and Texture-driven Generative Adversarial Network for Lung Nodule Segmentation
Abstract Slides Poster Similar
SA-UNet: Spatial Attention U-Net for Retinal Vessel Segmentation
Changlu Guo, Marton Szemenyei, Yugen Yi, Wenle Wang, Buer Chen, Changqi Fan

Auto-TLDR; Spatial Attention U-Net for Segmentation of Retinal Blood Vessels
Abstract Slides Poster Similar
Unsupervised Detection of Pulmonary Opacities for Computer-Aided Diagnosis of COVID-19 on CT Images
Rui Xu, Xiao Cao, Yufeng Wang, Yen-Wei Chen, Xinchen Ye, Lin Lin, Wenchao Zhu, Chao Chen, Fangyi Xu, Yong Zhou, Hongjie Hu, Shoji Kido, Noriyuki Tomiyama
Auto-TLDR; A computer-aided diagnosis of COVID-19 from CT images using unsupervised pulmonary opacity detection
Abstract Slides Poster Similar
A Lumen Segmentation Method in Ureteroscopy Images Based on a Deep Residual U-Net Architecture
Jorge Lazo, Marzullo Aldo, Sara Moccia, Michele Catellani, Benoit Rosa, Elena De Momi, Michel De Mathelin, Francesco Calimeri
Auto-TLDR; A Deep Neural Network for Ureteroscopy with Residual Units
Abstract Slides Poster Similar
Deep Recurrent-Convolutional Model for AutomatedSegmentation of Craniomaxillofacial CT Scans
Francesca Murabito, Simone Palazzo, Federica Salanitri Proietto, Francesco Rundo, Ulas Bagci, Daniela Giordano, Rosalia Leonardi, Concetto Spampinato
Auto-TLDR; Automated Segmentation of Anatomical Structures in Craniomaxillofacial CT Scans using Fully Convolutional Deep Networks
Abstract Slides Poster Similar
Learning Defects in Old Movies from Manually Assisted Restoration
Arthur Renaudeau, Travis Seng, Axel Carlier, Jean-Denis Durou, Fabien Pierre, Francois Lauze, Jean-François Aujol

Auto-TLDR; U-Net: Detecting Defects in Old Movies by Inpainting Techniques
Abstract Slides Poster Similar
A New Geodesic-Based Feature for Characterization of 3D Shapes: Application to Soft Tissue Organ Temporal Deformations
Karim Makki, Amine Bohi, Augustin Ogier, Marc-Emmanuel Bellemare

Auto-TLDR; Spatio-Temporal Feature Descriptors for 3D Shape Characterization from Point Clouds
Abstract Slides Poster Similar
Deep Learning-Based Type Identification of Volumetric MRI Sequences
Jean Pablo De Mello, Thiago Paixão, Rodrigo Berriel, Mauricio Reyes, Alberto F. De Souza, Claudine Badue, Thiago Oliveira-Santos

Auto-TLDR; Deep Learning for Brain MRI Sequences Identification Using Convolutional Neural Network
Abstract Slides Poster Similar
Few Shot Learning Framework to Reduce Inter-Observer Variability in Medical Images

Auto-TLDR; Few-Shot Learning for Quality Image Annotation
Abstract Slides Poster Similar
Robust Skeletonization for Plant Root Structure Reconstruction from MRI

Auto-TLDR; Structural reconstruction of plant roots from MRI using semantic root vs shoot segmentation and 3D skeletonization
Abstract Slides Poster Similar
Force Banner for the Recognition of Spatial Relations
Robin Deléarde, Camille Kurtz, Laurent Wendling, Philippe Dejean

Auto-TLDR; Spatial Relation Recognition using Force Banners
Weakly Supervised Geodesic Segmentation of Egyptian Mummy CT Scans
Avik Hati, Matteo Bustreo, Diego Sona, Vittorio Murino, Alessio Del Bue

Auto-TLDR; A Weakly Supervised and Efficient Interactive Segmentation of Ancient Egyptian Mummies CT Scans Using Geodesic Distance Measure and GrabCut
Abstract Slides Poster Similar
Segmenting Kidney on Multiple Phase CT Images Using ULBNet
Yanling Chi, Yuyu Xu, Gang Feng, Jiawei Mao, Sihua Wu, Guibin Xu, Weimin Huang
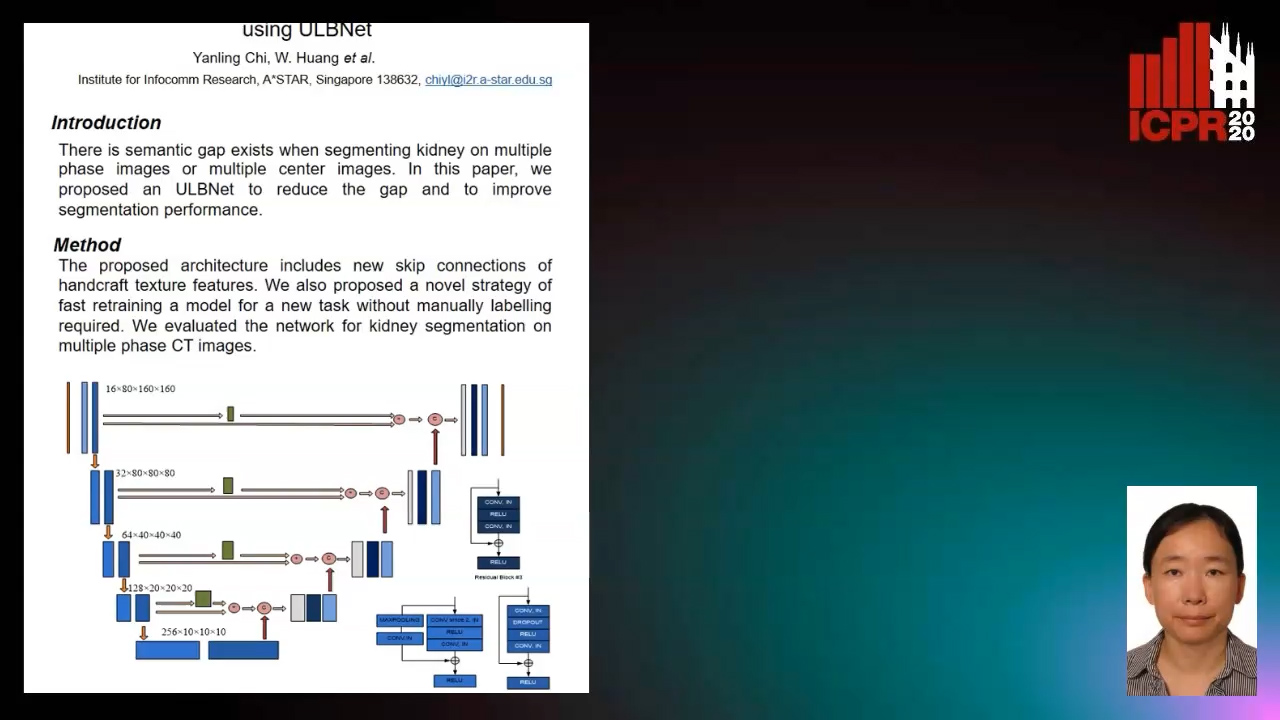
Auto-TLDR; A ULBNet network for kidney segmentation on multiple phase CT images
Evaluation of Anomaly Detection Algorithms for the Real-World Applications
Marija Ivanovska, Domen Tabernik, Danijel Skocaj, Janez Pers

Auto-TLDR; Evaluating Anomaly Detection Algorithms for Practical Applications
Abstract Slides Poster Similar
Automatic Tuberculosis Detection Using Chest X-Ray Analysis with Position Enhanced Structural Information
Hermann Jepdjio Nkouanga, Szilard Vajda
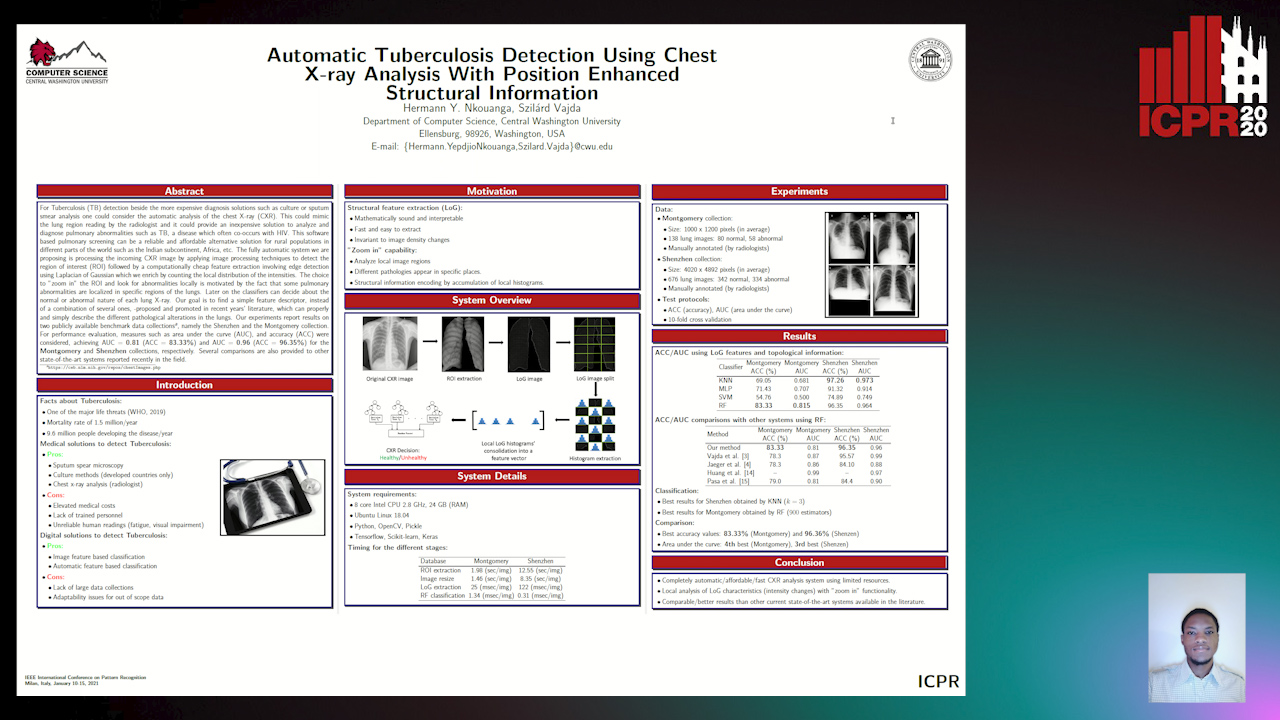
Auto-TLDR; Automatic Chest X-ray Screening for Tuberculosis in Rural Population using Localized Region on Interest
Abstract Slides Poster Similar
Offset Curves Loss for Imbalanced Problem in Medical Segmentation
Ngan Le, Duc Toan Bui, Khoa Luu, Marios Savvides

Auto-TLDR; Offset Curves Loss for Medical Image Segmentation
Deep Multi-Stage Model for Automated Landmarking of Craniomaxillofacial CT Scans
Simone Palazzo, Giovanni Bellitto, Luca Prezzavento, Francesco Rundo, Ulas Bagci, Daniela Giordano, Rosalia Leonardi, Concetto Spampinato

Auto-TLDR; Automated Landmarking of Craniomaxillofacial CT Images Using Deep Multi-Stage Architecture
One Step Clustering Based on A-Contrario Framework for Detection of Alterations in Historical Violins
Alireza Rezaei, Sylvie Le Hégarat-Mascle, Emanuel Aldea, Piercarlo Dondi, Marco Malagodi

Auto-TLDR; A-Contrario Clustering for the Detection of Altered Violins using UVIFL Images
Abstract Slides Poster Similar
Fine-Tuning Convolutional Neural Networks: A Comprehensive Guide and Benchmark Analysis for Glaucoma Screening
Amed Mvoulana, Rostom Kachouri, Mohamed Akil
Auto-TLDR; Fine-tuning Convolutional Neural Networks for Glaucoma Screening
Abstract Slides Poster Similar
End-To-End Multi-Task Learning for Lung Nodule Segmentation and Diagnosis
Wei Chen, Qiuli Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; A novel multi-task framework for lung nodule diagnosis based on deep learning and medical features
On Morphological Hierarchies for Image Sequences
Caglayan Tuna, Alain Giros, François Merciol, Sébastien Lefèvre

Auto-TLDR; Comparison of Hierarchies for Image Sequences
Abstract Slides Poster Similar
MedZip: 3D Medical Images Lossless Compressor Using Recurrent Neural Network (LSTM)
Omniah Nagoor, Joss Whittle, Jingjing Deng, Benjamin Mora, Mark W. Jones
Auto-TLDR; Recurrent Neural Network for Lossless Medical Image Compression using Long Short-Term Memory
Graph-Based Image Decoding for Multiplexed in Situ RNA Detection
Gabriele Partel, Carolina Wahlby
Auto-TLDR; A Graph-based Decoding Approach for Multiplexed In situ RNA Detection
Using Machine Learning to Refer Patients with Chronic Kidney Disease to Secondary Care
Lee Au-Yeung, Xianghua Xie, Timothy Marcus Scale, James Anthony Chess

Auto-TLDR; A Machine Learning Approach for Chronic Kidney Disease Prediction using Blood Test Data
Abstract Slides Poster Similar
One-Stage Multi-Task Detector for 3D Cardiac MR Imaging
Weizeng Lu, Xi Jia, Wei Chen, Nicolò Savioli, Antonio De Marvao, Linlin Shen, Declan O'Regan, Jinming Duan

Auto-TLDR; Multi-task Learning for Real-Time, simultaneous landmark location and bounding box detection in 3D space
Abstract Slides Poster Similar
Prediction of Obstructive Coronary Artery Disease from Myocardial Perfusion Scintigraphy using Deep Neural Networks
Ida Arvidsson, Niels Christian Overgaard, Miguel Ochoa Figueroa, Jeronimo Rose, Anette Davidsson, Kalle Åström, Anders Heyden

Auto-TLDR; A Deep Learning Algorithm for Multi-label Classification of Myocardial Perfusion Scintigraphy for Stable Ischemic Heart Disease
Abstract Slides Poster Similar
BG-Net: Boundary-Guided Network for Lung Segmentation on Clinical CT Images
Rui Xu, Yi Wang, Tiantian Liu, Xinchen Ye, Lin Lin, Yen-Wei Chen, Shoji Kido, Noriyuki Tomiyama
Auto-TLDR; Boundary-Guided Network for Lung Segmentation on CT Images
Abstract Slides Poster Similar
Extended Depth of Field Preserving Color Fidelity for Automated Digital Cytology
Alexandre Bouyssoux, Riadh Fezzani, Jean-Christophe Olivo-Marin
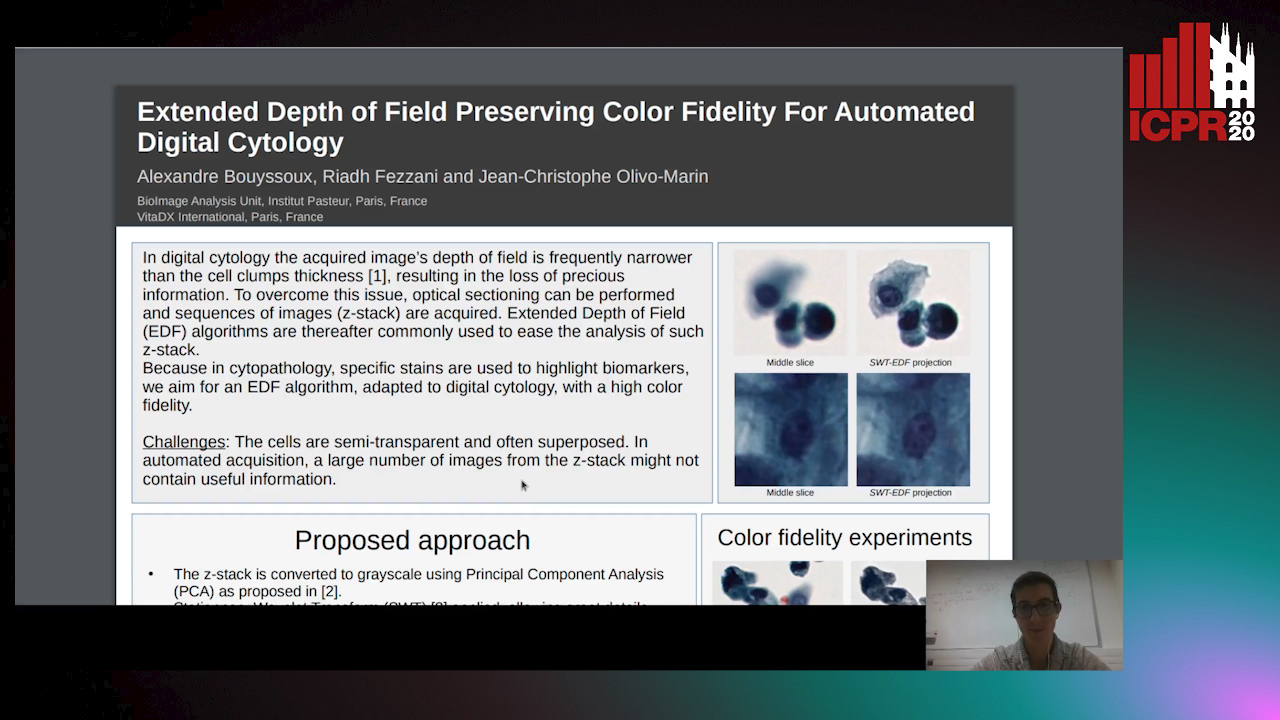
Auto-TLDR; Multi-Channel Extended Depth of Field for Digital cytology based on the stationary wavelet transform
Leveraging Unlabeled Data for Glioma Molecular Subtype and Survival Prediction
Nicholas Nuechterlein, Beibin Li, Mehmet Saygin Seyfioglu, Sachin Mehta, Patrick Cimino, Linda Shapiro

Auto-TLDR; Multimodal Brain Tumor Segmentation Using Unlabeled MR Data and Genomic Data for Cancer Prediction
Abstract Slides Poster Similar