A Novel Computer-Aided Diagnostic System for Early Assessment of Hepatocellular Carcinoma
Ahmed Alksas,
Mohamed Shehata,
Gehad Saleh,
Ahmed Shaffie,
Ahmed Soliman,
Mohammed Ghazal,
Hadil Abukhalifeh,
Abdel Razek Ahmed,
Ayman El-Baz

Auto-TLDR; Classification of Liver Tumor Lesions from CE-MRI Using Structured Structural Features and Functional Features
Similar papers
A Benchmark Dataset for Segmenting Liver, Vasculature and Lesions from Large-Scale Computed Tomography Data
Bo Wang, Zhengqing Xu, Wei Xu, Qingsen Yan, Liang Zhang, Zheng You

Auto-TLDR; The Biggest Treatment-Oriented Liver Cancer Dataset for Segmentation
Abstract Slides Poster Similar
A Comparison of Neural Network Approaches for Melanoma Classification
Maria Frasca, Michele Nappi, Michele Risi, Genoveffa Tortora, Alessia Auriemma Citarella
Auto-TLDR; Classification of Melanoma Using Deep Neural Network Methodologies
Abstract Slides Poster Similar
Segmentation of Axillary and Supraclavicular Tumoral Lymph Nodes in PET/CT: A Hybrid CNN/Component-Tree Approach
Diana Lucia Farfan Cabrera, Nicolas Gogin, David Morland, Benoît Naegel, Dimitri Papathanassiou, Nicolas Passat

Auto-TLDR; Coupling Convolutional Neural Networks and Component-Trees for Lymph node Segmentation from PET/CT Images
Automatic Classification of Human Granulosa Cells in Assisted Reproductive Technology Using Vibrational Spectroscopy Imaging
Marina Paolanti, Emanuele Frontoni, Giorgia Gioacchini, Giorgini Elisabetta, Notarstefano Valentina, Zacà Carlotta, Carnevali Oliana, Andrea Borini, Marco Mameli

Auto-TLDR; Predicting Oocyte Quality in Assisted Reproductive Technology Using Machine Learning Techniques
Abstract Slides Poster Similar
Leveraging Unlabeled Data for Glioma Molecular Subtype and Survival Prediction
Nicholas Nuechterlein, Beibin Li, Mehmet Saygin Seyfioglu, Sachin Mehta, Patrick Cimino, Linda Shapiro

Auto-TLDR; Multimodal Brain Tumor Segmentation Using Unlabeled MR Data and Genomic Data for Cancer Prediction
Abstract Slides Poster Similar
Unsupervised Detection of Pulmonary Opacities for Computer-Aided Diagnosis of COVID-19 on CT Images
Rui Xu, Xiao Cao, Yufeng Wang, Yen-Wei Chen, Xinchen Ye, Lin Lin, Wenchao Zhu, Chao Chen, Fangyi Xu, Yong Zhou, Hongjie Hu, Shoji Kido, Noriyuki Tomiyama
Auto-TLDR; A computer-aided diagnosis of COVID-19 from CT images using unsupervised pulmonary opacity detection
Abstract Slides Poster Similar
Neural Machine Registration for Motion Correction in Breast DCE-MRI
Federica Aprea, Stefano Marrone, Carlo Sansone
Auto-TLDR; A Neural Registration Network for Dynamic Contrast Enhanced-Magnetic Resonance Imaging
Abstract Slides Poster Similar
Automatic Tuberculosis Detection Using Chest X-Ray Analysis with Position Enhanced Structural Information
Hermann Jepdjio Nkouanga, Szilard Vajda
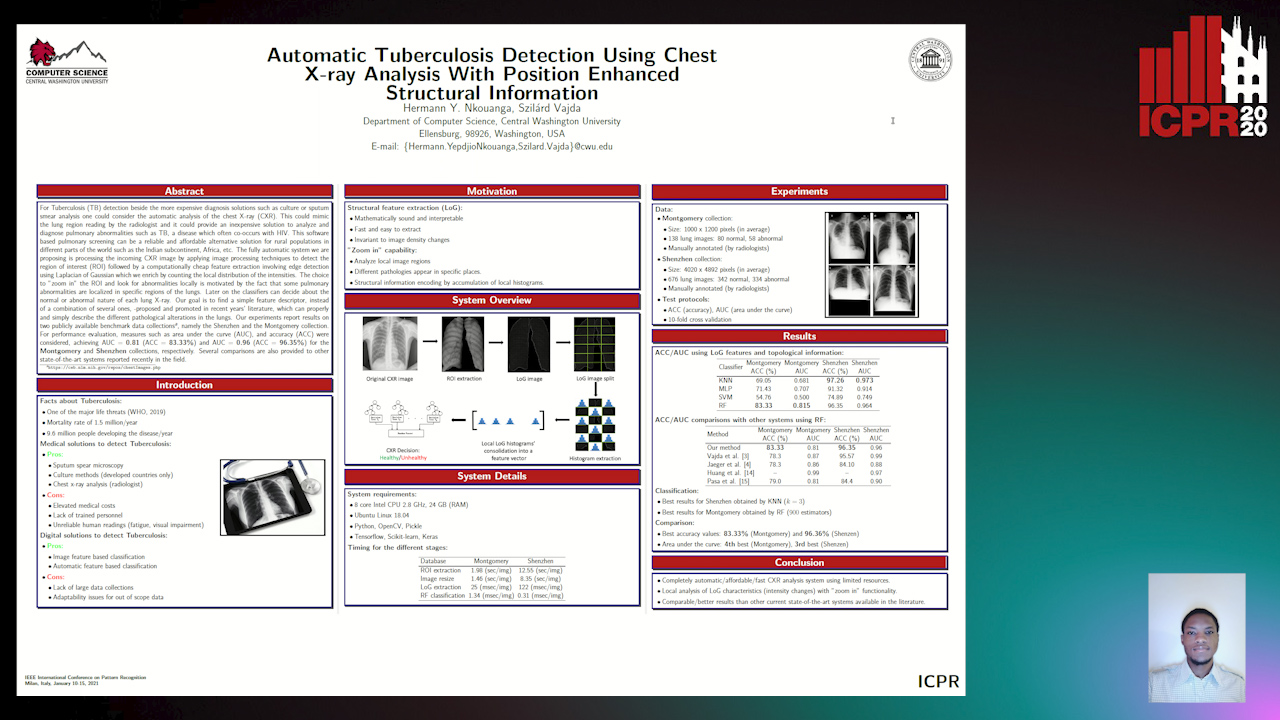
Auto-TLDR; Automatic Chest X-ray Screening for Tuberculosis in Rural Population using Localized Region on Interest
Abstract Slides Poster Similar
Automatic Semantic Segmentation of Structural Elements related to the Spinal Cord in the Lumbar Region by Using Convolutional Neural Networks
Jhon Jairo Sáenz Gamboa, Maria De La Iglesia-Vaya, Jon Ander Gómez

Auto-TLDR; Semantic Segmentation of Lumbar Spine Using Convolutional Neural Networks
Abstract Slides Poster Similar
Classify Breast Histopathology Images with Ductal Instance-Oriented Pipeline
Beibin Li, Ezgi Mercan, Sachin Mehta, Stevan Knezevich, Corey Arnold, Donald Weaver, Joann Elmore, Linda Shapiro

Auto-TLDR; DIOP: Ductal Instance-Oriented Pipeline for Diagnostic Classification
Abstract Slides Poster Similar
Vesselness Filters: A Survey with Benchmarks Applied to Liver Imaging
Jonas Lamy, Odyssée Merveille, Bertrand Kerautret, Nicolas Passat, Antoine Vacavant

Auto-TLDR; Comparison of Vessel Enhancement Filters for Liver Vascular Network Segmentation
Abstract Slides Poster Similar
A Systematic Investigation on Deep Architectures for Automatic Skin Lesions Classification
Pierluigi Carcagni, Marco Leo, Andrea Cuna, Giuseppe Celeste, Cosimo Distante

Auto-TLDR; RegNet: Deep Investigation of Convolutional Neural Networks for Automatic Classification of Skin Lesions
Abstract Slides Poster Similar
Tensor Factorization of Brain Structural Graph for Unsupervised Classification in Multiple Sclerosis
Berardino Barile, Marzullo Aldo, Claudio Stamile, Françoise Durand-Dubief, Dominique Sappey-Marinier
Auto-TLDR; A Fully Automated Tensor-based Algorithm for Multiple Sclerosis Classification based on Structural Connectivity Graph of the White Matter Network
Abstract Slides Poster Similar
Supporting Skin Lesion Diagnosis with Content-Based Image Retrieval
Stefano Allegretti, Federico Bolelli, Federico Pollastri, Sabrina Longhitano, Giovanni Pellacani, Costantino Grana

Auto-TLDR; Skin Images Retrieval Using Convolutional Neural Networks for Skin Lesion Classification and Segmentation
Abstract Slides Poster Similar
Prediction of Obstructive Coronary Artery Disease from Myocardial Perfusion Scintigraphy using Deep Neural Networks
Ida Arvidsson, Niels Christian Overgaard, Miguel Ochoa Figueroa, Jeronimo Rose, Anette Davidsson, Kalle Åström, Anders Heyden

Auto-TLDR; A Deep Learning Algorithm for Multi-label Classification of Myocardial Perfusion Scintigraphy for Stable Ischemic Heart Disease
Abstract Slides Poster Similar
Planar 3D Transfer Learning for End to End Unimodal MRI Unbalanced Data Segmentation
Martin Kolarik, Radim Burget, Carlos M. Travieso-Gonzalez, Jan Kocica

Auto-TLDR; Planar 3D Res-U-Net Network for Unbalanced 3D Image Segmentation using Fluid Attenuation Inversion Recover
Deep Learning-Based Type Identification of Volumetric MRI Sequences
Jean Pablo De Mello, Thiago Paixão, Rodrigo Berriel, Mauricio Reyes, Alberto F. De Souza, Claudine Badue, Thiago Oliveira-Santos

Auto-TLDR; Deep Learning for Brain MRI Sequences Identification Using Convolutional Neural Network
Abstract Slides Poster Similar
Using Machine Learning to Refer Patients with Chronic Kidney Disease to Secondary Care
Lee Au-Yeung, Xianghua Xie, Timothy Marcus Scale, James Anthony Chess

Auto-TLDR; A Machine Learning Approach for Chronic Kidney Disease Prediction using Blood Test Data
Abstract Slides Poster Similar
SAGE: Sequential Attribute Generator for Analyzing Glioblastomas Using Limited Dataset
Padmaja Jonnalagedda, Brent Weinberg, Jason Allen, Taejin Min, Shiv Bhanu, Bir Bhanu

Auto-TLDR; SAGE: Generative Adversarial Networks for Imaging Biomarker Detection and Prediction
Abstract Slides Poster Similar
3D Medical Multi-Modal Segmentation Network Guided by Multi-Source Correlation Constraint
Tongxue Zhou, Stéphane Canu, Pierre Vera, Su Ruan
Auto-TLDR; Multi-modality Segmentation with Correlation Constrained Network
Abstract Slides Poster Similar
A Multi-Task Contextual Atrous Residual Network for Brain Tumor Detection & Segmentation
Ngan Le, Kashu Yamazaki, Quach Kha Gia, Thanh-Dat Truong, Marios Savvides

Auto-TLDR; Contextual Brain Tumor Segmentation Using 3D atrous Residual Networks and Cascaded Structures
DARN: Deep Attentive Refinement Network for Liver Tumor Segmentation from 3D CT Volume
Yao Zhang, Jiang Tian, Cheng Zhong, Yang Zhang, Zhongchao Shi, Zhiqiang He

Auto-TLDR; Deep Attentive Refinement Network for Liver Tumor Segmentation from 3D Computed Tomography Using Multi-Level Features
Abstract Slides Poster Similar
A Deep Learning Approach for the Segmentation of Myocardial Diseases
Khawala Brahim, Abdull Qayyum, Alain Lalande, Arnaud Boucher, Anis Sakly, Fabrice Meriaudeau
Auto-TLDR; Segmentation of Myocardium Infarction Using Late GADEMRI and SegU-Net
Abstract Slides Poster Similar
Encoding Brain Networks through Geodesic Clustering of Functional Connectivity for Multiple Sclerosis Classification
Muhammad Abubakar Yamin, Valsasina Paola, Michael Dayan, Sebastiano Vascon, Tessadori Jacopo, Filippi Massimo, Vittorio Murino, A Rocca Maria, Diego Sona

Auto-TLDR; Geodesic Clustering of Connectivity Matrices for Multiple Sclerosis Classification
Abstract Slides Poster Similar
Inferring Functional Properties from Fluid Dynamics Features
Andrea Schillaci, Maurizio Quadrio, Carlotta Pipolo, Marcello Restelli, Giacomo Boracchi

Auto-TLDR; Exploiting Convective Properties of Computational Fluid Dynamics for Medical Diagnosis
Abstract Slides Poster Similar
Dealing with Scarce Labelled Data: Semi-Supervised Deep Learning with Mix Match for Covid-19 Detection Using Chest X-Ray Images
Saúl Calderón Ramirez, Raghvendra Giri, Shengxiang Yang, Armaghan Moemeni, Mario Umaña, David Elizondo, Jordina Torrents-Barrena, Miguel A. Molina-Cabello

Auto-TLDR; Semi-supervised Deep Learning for Covid-19 Detection using Chest X-rays
Abstract Slides Poster Similar
A Deep Learning-Based Method for Predicting Volumes of Nasopharyngeal Carcinoma for Adaptive Radiation Therapy Treatment
Bilel Daoud, Ken'Ichi Morooka, Shoko Miyauchi, Ryo Kurazume, Wafa Mnejja, Leila Farhat, Jamel Daoud

Auto-TLDR; TEP-Net: Tumor Evolution Prediction of Nasopharyngeal Carcinoma and Organ-at-risks Using CT Images
Abstract Slides Poster Similar
MTGAN: Mask and Texture-Driven Generative Adversarial Network for Lung Nodule Segmentation
Wei Chen, Qiuli Wang, Kun Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; Mask and Texture-driven Generative Adversarial Network for Lung Nodule Segmentation
Abstract Slides Poster Similar
Inception Based Deep Learning Architecture for Tuberculosis Screening of Chest X-Rays
Dipayan Das, K.C. Santosh, Umapada Pal

Auto-TLDR; End to End CNN-based Chest X-ray Screening for Tuberculosis positive patients in the severely resource constrained regions of the world
Abstract Slides Poster Similar
A Lumen Segmentation Method in Ureteroscopy Images Based on a Deep Residual U-Net Architecture
Jorge Lazo, Marzullo Aldo, Sara Moccia, Michele Catellani, Benoit Rosa, Elena De Momi, Michel De Mathelin, Francesco Calimeri
Auto-TLDR; A Deep Neural Network for Ureteroscopy with Residual Units
Abstract Slides Poster Similar
A New Geodesic-Based Feature for Characterization of 3D Shapes: Application to Soft Tissue Organ Temporal Deformations
Karim Makki, Amine Bohi, Augustin Ogier, Marc-Emmanuel Bellemare

Auto-TLDR; Spatio-Temporal Feature Descriptors for 3D Shape Characterization from Point Clouds
Abstract Slides Poster Similar
Breast Anatomy Enriched Tumor Saliency Estimation
Fei Xu, Yingtao Zhang, Heng-Da Cheng, Jianrui Ding, Boyu Zhang, Chunping Ning, Ying Wang

Auto-TLDR; Tumor Saliency Estimation for Breast Ultrasound using enriched breast anatomy knowledge
Abstract Slides Poster Similar
Trainable Spectrally Initializable Matrix Transformations in Convolutional Neural Networks
Michele Alberti, Angela Botros, Schuetz Narayan, Rolf Ingold, Marcus Liwicki, Mathias Seuret
Auto-TLDR; Trainable and Spectrally Initializable Matrix Transformations for Neural Networks
Abstract Slides Poster Similar
Deep Multi-Stage Model for Automated Landmarking of Craniomaxillofacial CT Scans
Simone Palazzo, Giovanni Bellitto, Luca Prezzavento, Francesco Rundo, Ulas Bagci, Daniela Giordano, Rosalia Leonardi, Concetto Spampinato

Auto-TLDR; Automated Landmarking of Craniomaxillofacial CT Images Using Deep Multi-Stage Architecture
Deep Transfer Learning for Alzheimer’s Disease Detection
Nicole Cilia, Claudio De Stefano, Francesco Fontanella, Claudio Marrocco, Mario Molinara, Alessandra Scotto Di Freca

Auto-TLDR; Automatic Detection of Handwriting Alterations for Alzheimer's Disease Diagnosis using Dynamic Features
Abstract Slides Poster Similar
End-To-End Multi-Task Learning for Lung Nodule Segmentation and Diagnosis
Wei Chen, Qiuli Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; A novel multi-task framework for lung nodule diagnosis based on deep learning and medical features
A Heuristic-Based Decision Tree for Connected Components Labeling of 3D Volumes
Maximilian Söchting, Stefano Allegretti, Federico Bolelli, Costantino Grana

Auto-TLDR; Entropy Partitioning Decision Tree for Connected Components Labeling
Abstract Slides Poster Similar
Investigating and Exploiting Image Resolution for Transfer Learning-Based Skin Lesion Classification
Amirreza Mahbod, Gerald Schaefer, Chunliang Wang, Rupert Ecker, Georg Dorffner, Isabella Ellinger

Auto-TLDR; Fine-tuned Neural Networks for Skin Lesion Classification Using Dermoscopic Images
Abstract Slides Poster Similar
Longitudinal Feature Selection and Feature Learning for Parkinson’s Disease Diagnosis and Prediction
Haijun Lei, Zhongwei Huang, Xiaohua Xiao, Yi Lei, En-Leng Tan, Baiying Lei, Shiqi Li

Auto-TLDR; Joint Learning from Multiple Modalities and Relations for Joint Disease Diagnosis and Prediction in Parkinson's Disease
Abstract Slides Poster Similar
A Riemannian Framework for Detecting Stimulus-Relevant Fiber Pathways
Jingyong Su, Linlin Tang, Zhipeng Yang, Mengmeng Guo

Auto-TLDR; Clustering Task-Specific Fiber Pathways in Functional MRI using BOLD Signals
Confidence Calibration for Deep Renal Biopsy Immunofluorescence Image Classification
Federico Pollastri, Juan Maroñas, Federico Bolelli, Giulia Ligabue, Roberto Paredes, Riccardo Magistroni, Costantino Grana

Auto-TLDR; A Probabilistic Convolutional Neural Network for Immunofluorescence Classification in Renal Biopsy
Abstract Slides Poster Similar
A General End-To-End Method for Characterizing Neuropsychiatric Disorders Using Free-Viewing Visual Scanning Tasks
Hong Yue Sean Liu, Jonathan Chung, Moshe Eizenman

Auto-TLDR; A general, data-driven, end-to-end framework that extracts relevant features of attentional bias from visual scanning behaviour and uses these features
Abstract Slides Poster Similar
Influence of Event Duration on Automatic Wheeze Classification
Bruno M Rocha, Diogo Pessoa, Alda Marques, Paulo Carvalho, Rui Pedro Paiva

Auto-TLDR; Experimental Design of the Non-wheeze Class for Wheeze Classification
Abstract Slides Poster Similar
Segmenting Kidney on Multiple Phase CT Images Using ULBNet
Yanling Chi, Yuyu Xu, Gang Feng, Jiawei Mao, Sihua Wu, Guibin Xu, Weimin Huang
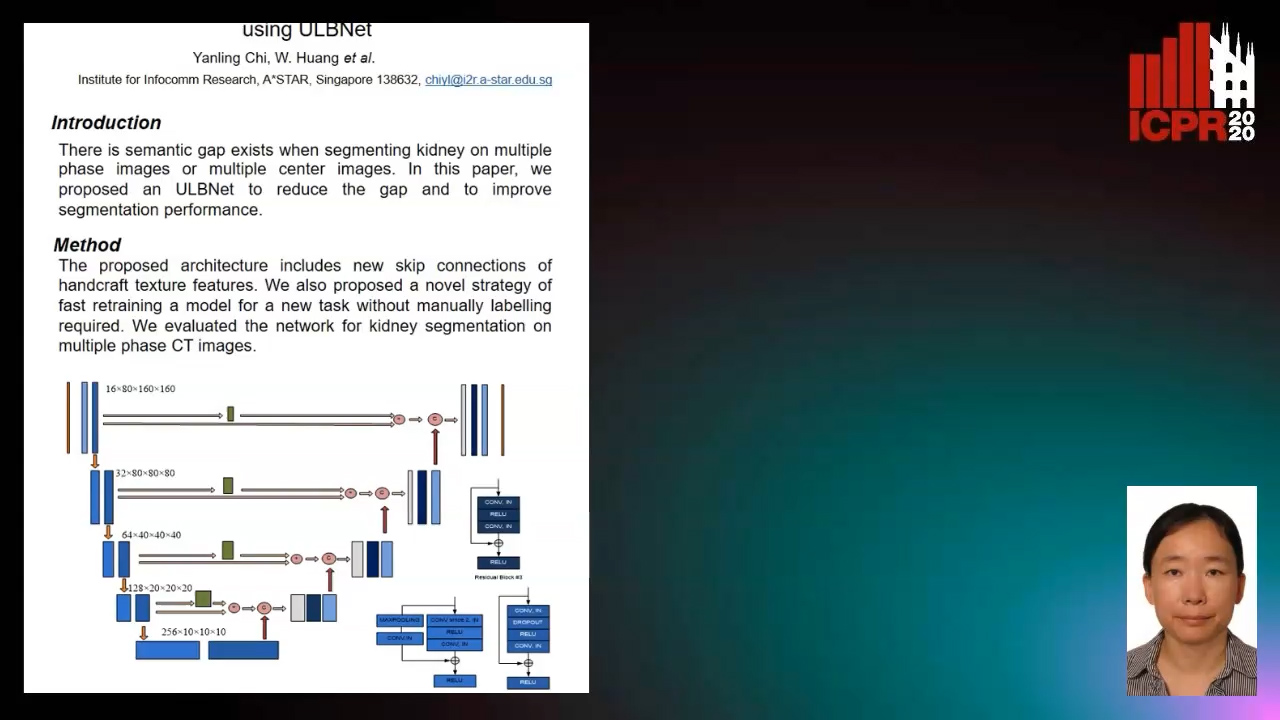
Auto-TLDR; A ULBNet network for kidney segmentation on multiple phase CT images
Fused 3-Stage Image Segmentation for Pleural Effusion Cell Clusters
Sike Ma, Meng Zhao, Hao Wang, Fan Shi, Xuguo Sun, Shengyong Chen, Hong-Ning Dai

Auto-TLDR; Coarse Segmentation of Stained and Stained Unstained Cell Clusters in pleural effusion using 3-stage segmentation method
Abstract Slides Poster Similar
Semantic Segmentation of Breast Ultrasound Image with Pyramid Fuzzy Uncertainty Reduction and Direction Connectedness Feature
Kuan Huang, Yingtao Zhang, Heng-Da Cheng, Ping Xing, Boyu Zhang

Auto-TLDR; Uncertainty-Based Deep Learning for Breast Ultrasound Image Segmentation
Abstract Slides Poster Similar
Deep Learning Based Sepsis Intervention: The Modelling and Prediction of Severe Sepsis Onset

Auto-TLDR; Predicting Sepsis onset by up to six hours prior using a boosted cascading training methodology and adjustable margin hinge loss function
Abstract Slides Poster Similar
Deep Recurrent-Convolutional Model for AutomatedSegmentation of Craniomaxillofacial CT Scans
Francesca Murabito, Simone Palazzo, Federica Salanitri Proietto, Francesco Rundo, Ulas Bagci, Daniela Giordano, Rosalia Leonardi, Concetto Spampinato
Auto-TLDR; Automated Segmentation of Anatomical Structures in Craniomaxillofacial CT Scans using Fully Convolutional Deep Networks
Abstract Slides Poster Similar