Segmentation of Axillary and Supraclavicular Tumoral Lymph Nodes in PET/CT: A Hybrid CNN/Component-Tree Approach
Diana Lucia Farfan Cabrera,
Nicolas Gogin,
David Morland,
Benoît Naegel,
Dimitri Papathanassiou,
Nicolas Passat

Auto-TLDR; Coupling Convolutional Neural Networks and Component-Trees for Lymph node Segmentation from PET/CT Images
Similar papers
Vesselness Filters: A Survey with Benchmarks Applied to Liver Imaging
Jonas Lamy, Odyssée Merveille, Bertrand Kerautret, Nicolas Passat, Antoine Vacavant

Auto-TLDR; Comparison of Vessel Enhancement Filters for Liver Vascular Network Segmentation
Abstract Slides Poster Similar
A Benchmark Dataset for Segmenting Liver, Vasculature and Lesions from Large-Scale Computed Tomography Data
Bo Wang, Zhengqing Xu, Wei Xu, Qingsen Yan, Liang Zhang, Zheng You

Auto-TLDR; The Biggest Treatment-Oriented Liver Cancer Dataset for Segmentation
Abstract Slides Poster Similar
A Deep Learning-Based Method for Predicting Volumes of Nasopharyngeal Carcinoma for Adaptive Radiation Therapy Treatment
Bilel Daoud, Ken'Ichi Morooka, Shoko Miyauchi, Ryo Kurazume, Wafa Mnejja, Leila Farhat, Jamel Daoud

Auto-TLDR; TEP-Net: Tumor Evolution Prediction of Nasopharyngeal Carcinoma and Organ-at-risks Using CT Images
Abstract Slides Poster Similar
MTGAN: Mask and Texture-Driven Generative Adversarial Network for Lung Nodule Segmentation
Wei Chen, Qiuli Wang, Kun Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; Mask and Texture-driven Generative Adversarial Network for Lung Nodule Segmentation
Abstract Slides Poster Similar
Deep Recurrent-Convolutional Model for AutomatedSegmentation of Craniomaxillofacial CT Scans
Francesca Murabito, Simone Palazzo, Federica Salanitri Proietto, Francesco Rundo, Ulas Bagci, Daniela Giordano, Rosalia Leonardi, Concetto Spampinato
Auto-TLDR; Automated Segmentation of Anatomical Structures in Craniomaxillofacial CT Scans using Fully Convolutional Deep Networks
Abstract Slides Poster Similar
A Novel Computer-Aided Diagnostic System for Early Assessment of Hepatocellular Carcinoma
Ahmed Alksas, Mohamed Shehata, Gehad Saleh, Ahmed Shaffie, Ahmed Soliman, Mohammed Ghazal, Hadil Abukhalifeh, Abdel Razek Ahmed, Ayman El-Baz

Auto-TLDR; Classification of Liver Tumor Lesions from CE-MRI Using Structured Structural Features and Functional Features
Abstract Slides Poster Similar
A Lumen Segmentation Method in Ureteroscopy Images Based on a Deep Residual U-Net Architecture
Jorge Lazo, Marzullo Aldo, Sara Moccia, Michele Catellani, Benoit Rosa, Elena De Momi, Michel De Mathelin, Francesco Calimeri
Auto-TLDR; A Deep Neural Network for Ureteroscopy with Residual Units
Abstract Slides Poster Similar
3D Medical Multi-Modal Segmentation Network Guided by Multi-Source Correlation Constraint
Tongxue Zhou, Stéphane Canu, Pierre Vera, Su Ruan
Auto-TLDR; Multi-modality Segmentation with Correlation Constrained Network
Abstract Slides Poster Similar
A Multi-Task Contextual Atrous Residual Network for Brain Tumor Detection & Segmentation
Ngan Le, Kashu Yamazaki, Quach Kha Gia, Thanh-Dat Truong, Marios Savvides

Auto-TLDR; Contextual Brain Tumor Segmentation Using 3D atrous Residual Networks and Cascaded Structures
BG-Net: Boundary-Guided Network for Lung Segmentation on Clinical CT Images
Rui Xu, Yi Wang, Tiantian Liu, Xinchen Ye, Lin Lin, Yen-Wei Chen, Shoji Kido, Noriyuki Tomiyama
Auto-TLDR; Boundary-Guided Network for Lung Segmentation on CT Images
Abstract Slides Poster Similar
DARN: Deep Attentive Refinement Network for Liver Tumor Segmentation from 3D CT Volume
Yao Zhang, Jiang Tian, Cheng Zhong, Yang Zhang, Zhongchao Shi, Zhiqiang He

Auto-TLDR; Deep Attentive Refinement Network for Liver Tumor Segmentation from 3D Computed Tomography Using Multi-Level Features
Abstract Slides Poster Similar
Unsupervised Detection of Pulmonary Opacities for Computer-Aided Diagnosis of COVID-19 on CT Images
Rui Xu, Xiao Cao, Yufeng Wang, Yen-Wei Chen, Xinchen Ye, Lin Lin, Wenchao Zhu, Chao Chen, Fangyi Xu, Yong Zhou, Hongjie Hu, Shoji Kido, Noriyuki Tomiyama
Auto-TLDR; A computer-aided diagnosis of COVID-19 from CT images using unsupervised pulmonary opacity detection
Abstract Slides Poster Similar
Automatic Semantic Segmentation of Structural Elements related to the Spinal Cord in the Lumbar Region by Using Convolutional Neural Networks
Jhon Jairo Sáenz Gamboa, Maria De La Iglesia-Vaya, Jon Ander Gómez

Auto-TLDR; Semantic Segmentation of Lumbar Spine Using Convolutional Neural Networks
Abstract Slides Poster Similar
Segmentation of Intracranial Aneurysm Remnant in MRA Using Dual-Attention Atrous Net
Subhashis Banerjee, Ashis Kumar Dhara, Johan Wikström, Robin Strand
Auto-TLDR; Dual-Attention Atrous Net for Segmentation of Intracranial Aneurysm Remnant from MRA Images
Abstract Slides Poster Similar
Deep Multi-Stage Model for Automated Landmarking of Craniomaxillofacial CT Scans
Simone Palazzo, Giovanni Bellitto, Luca Prezzavento, Francesco Rundo, Ulas Bagci, Daniela Giordano, Rosalia Leonardi, Concetto Spampinato

Auto-TLDR; Automated Landmarking of Craniomaxillofacial CT Images Using Deep Multi-Stage Architecture
Breast Anatomy Enriched Tumor Saliency Estimation
Fei Xu, Yingtao Zhang, Heng-Da Cheng, Jianrui Ding, Boyu Zhang, Chunping Ning, Ying Wang

Auto-TLDR; Tumor Saliency Estimation for Breast Ultrasound using enriched breast anatomy knowledge
Abstract Slides Poster Similar
Weakly Supervised Geodesic Segmentation of Egyptian Mummy CT Scans
Avik Hati, Matteo Bustreo, Diego Sona, Vittorio Murino, Alessio Del Bue

Auto-TLDR; A Weakly Supervised and Efficient Interactive Segmentation of Ancient Egyptian Mummies CT Scans Using Geodesic Distance Measure and GrabCut
Abstract Slides Poster Similar
A Deep Learning Approach for the Segmentation of Myocardial Diseases
Khawala Brahim, Abdull Qayyum, Alain Lalande, Arnaud Boucher, Anis Sakly, Fabrice Meriaudeau
Auto-TLDR; Segmentation of Myocardium Infarction Using Late GADEMRI and SegU-Net
Abstract Slides Poster Similar
Neural Machine Registration for Motion Correction in Breast DCE-MRI
Federica Aprea, Stefano Marrone, Carlo Sansone
Auto-TLDR; A Neural Registration Network for Dynamic Contrast Enhanced-Magnetic Resonance Imaging
Abstract Slides Poster Similar
A Comparison of Neural Network Approaches for Melanoma Classification
Maria Frasca, Michele Nappi, Michele Risi, Genoveffa Tortora, Alessia Auriemma Citarella
Auto-TLDR; Classification of Melanoma Using Deep Neural Network Methodologies
Abstract Slides Poster Similar
Planar 3D Transfer Learning for End to End Unimodal MRI Unbalanced Data Segmentation
Martin Kolarik, Radim Burget, Carlos M. Travieso-Gonzalez, Jan Kocica

Auto-TLDR; Planar 3D Res-U-Net Network for Unbalanced 3D Image Segmentation using Fluid Attenuation Inversion Recover
End-To-End Multi-Task Learning for Lung Nodule Segmentation and Diagnosis
Wei Chen, Qiuli Wang, Dan Yang, Xiaohong Zhang, Chen Liu, Yucong Li
Auto-TLDR; A novel multi-task framework for lung nodule diagnosis based on deep learning and medical features
FOANet: A Focus of Attention Network with Application to Myocardium Segmentation
Zhou Zhao, Elodie Puybareau, Nicolas Boutry, Thierry Geraud

Auto-TLDR; FOANet: A Hybrid Loss Function for Myocardium Segmentation of Cardiac Magnetic Resonance Images
Abstract Slides Poster Similar
A Transformer-Based Network for Anisotropic 3D Medical Image Segmentation
Guo Danfeng, Demetri Terzopoulos
Auto-TLDR; A transformer-based model to tackle the anisotropy problem in 3D medical image analysis
Abstract Slides Poster Similar
Segmenting Kidney on Multiple Phase CT Images Using ULBNet
Yanling Chi, Yuyu Xu, Gang Feng, Jiawei Mao, Sihua Wu, Guibin Xu, Weimin Huang
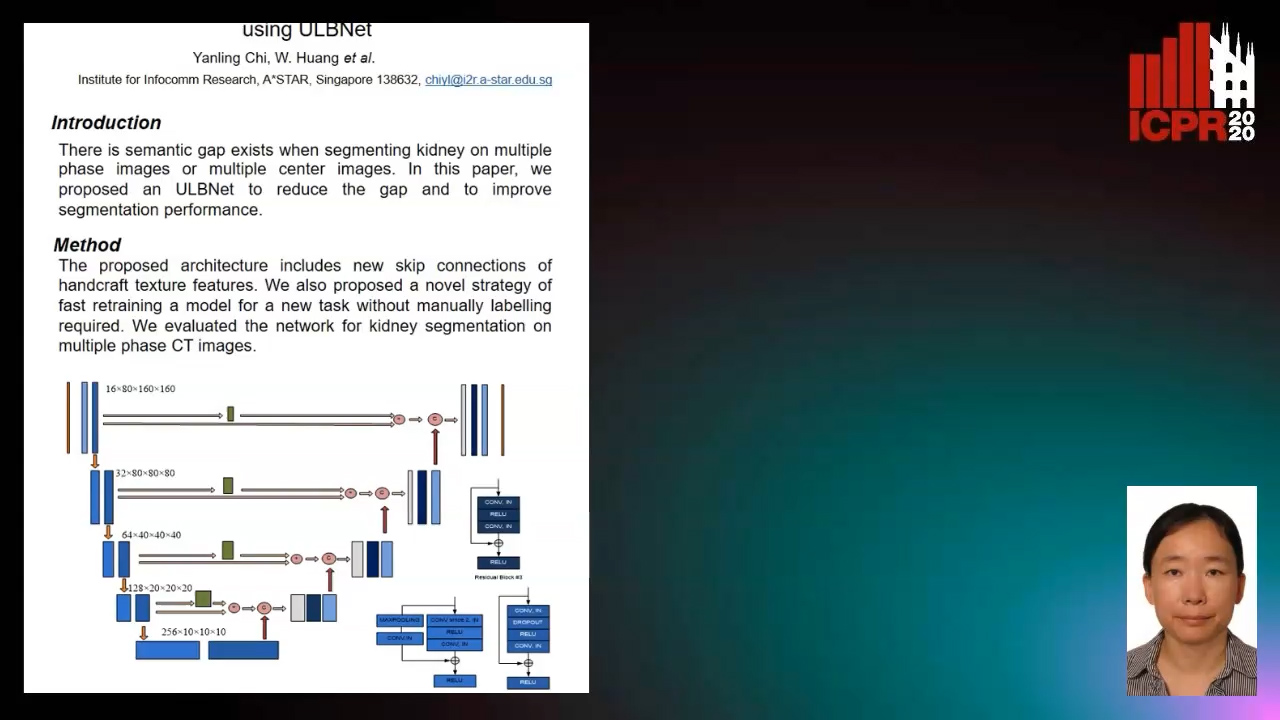
Auto-TLDR; A ULBNet network for kidney segmentation on multiple phase CT images
Leveraging Unlabeled Data for Glioma Molecular Subtype and Survival Prediction
Nicholas Nuechterlein, Beibin Li, Mehmet Saygin Seyfioglu, Sachin Mehta, Patrick Cimino, Linda Shapiro

Auto-TLDR; Multimodal Brain Tumor Segmentation Using Unlabeled MR Data and Genomic Data for Cancer Prediction
Abstract Slides Poster Similar
Learning to Segment Clustered Amoeboid Cells from Brightfield Microscopy Via Multi-Task Learning with Adaptive Weight Selection
Rituparna Sarkar, Suvadip Mukherjee, Elisabeth Labruyere, Jean-Christophe Olivo-Marin
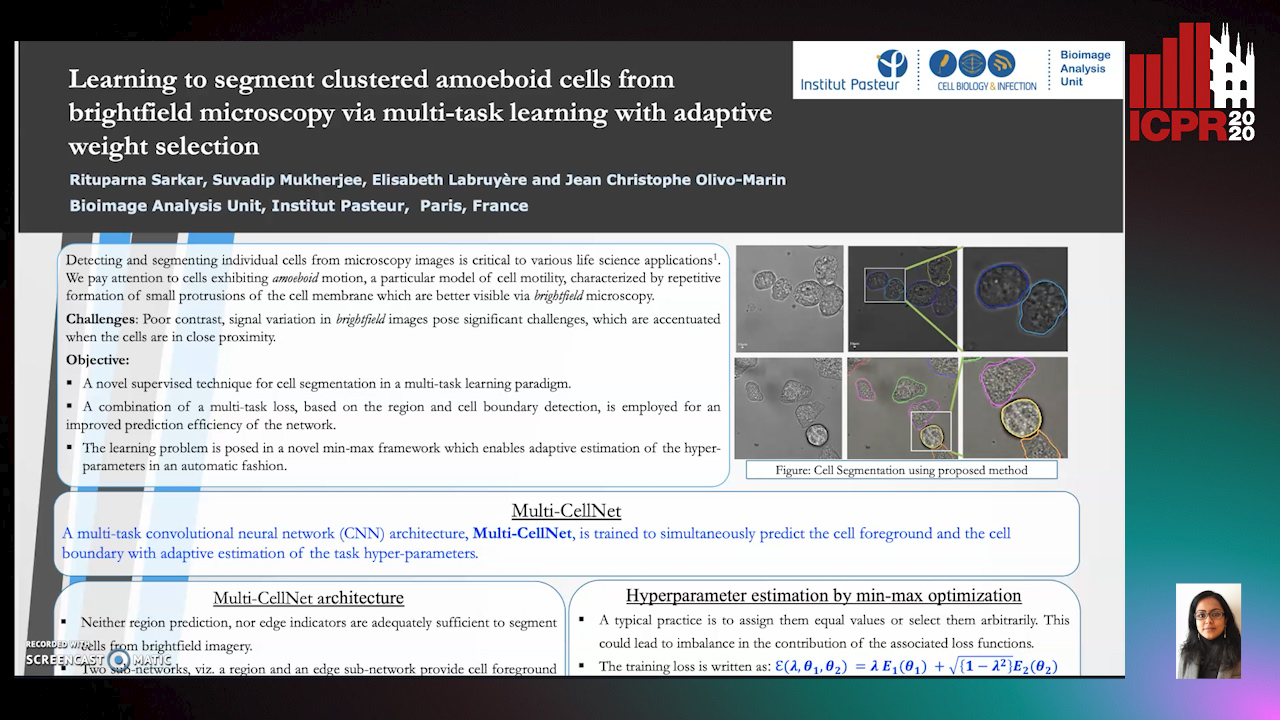
Auto-TLDR; Supervised Cell Segmentation from Microscopy Images using Multi-task Learning in a Multi-Task Learning Paradigm
BiLuNet: A Multi-Path Network for Semantic Segmentation on X-Ray Images
Van Luan Tran, Huei-Yung Lin, Rachel Liu, Chun-Han Tseng, Chun-Han Tseng
Auto-TLDR; BiLuNet: Multi-path Convolutional Neural Network for Semantic Segmentation of Lumbar vertebrae, sacrum,
Semantic Segmentation of Breast Ultrasound Image with Pyramid Fuzzy Uncertainty Reduction and Direction Connectedness Feature
Kuan Huang, Yingtao Zhang, Heng-Da Cheng, Ping Xing, Boyu Zhang

Auto-TLDR; Uncertainty-Based Deep Learning for Breast Ultrasound Image Segmentation
Abstract Slides Poster Similar
Robust Skeletonization for Plant Root Structure Reconstruction from MRI

Auto-TLDR; Structural reconstruction of plant roots from MRI using semantic root vs shoot segmentation and 3D skeletonization
Abstract Slides Poster Similar
DA-RefineNet: Dual-Inputs Attention RefineNet for Whole Slide Image Segmentation
Ziqiang Li, Rentuo Tao, Qianrun Wu, Bin Li

Auto-TLDR; DA-RefineNet: A dual-inputs attention network for whole slide image segmentation
Abstract Slides Poster Similar
Detecting Marine Species in Echograms Via Traditional, Hybrid, and Deep Learning Frameworks
Porto Marques Tunai, Alireza Rezvanifar, Melissa Cote, Alexandra Branzan Albu, Kaan Ersahin, Todd Mudge, Stephane Gauthier
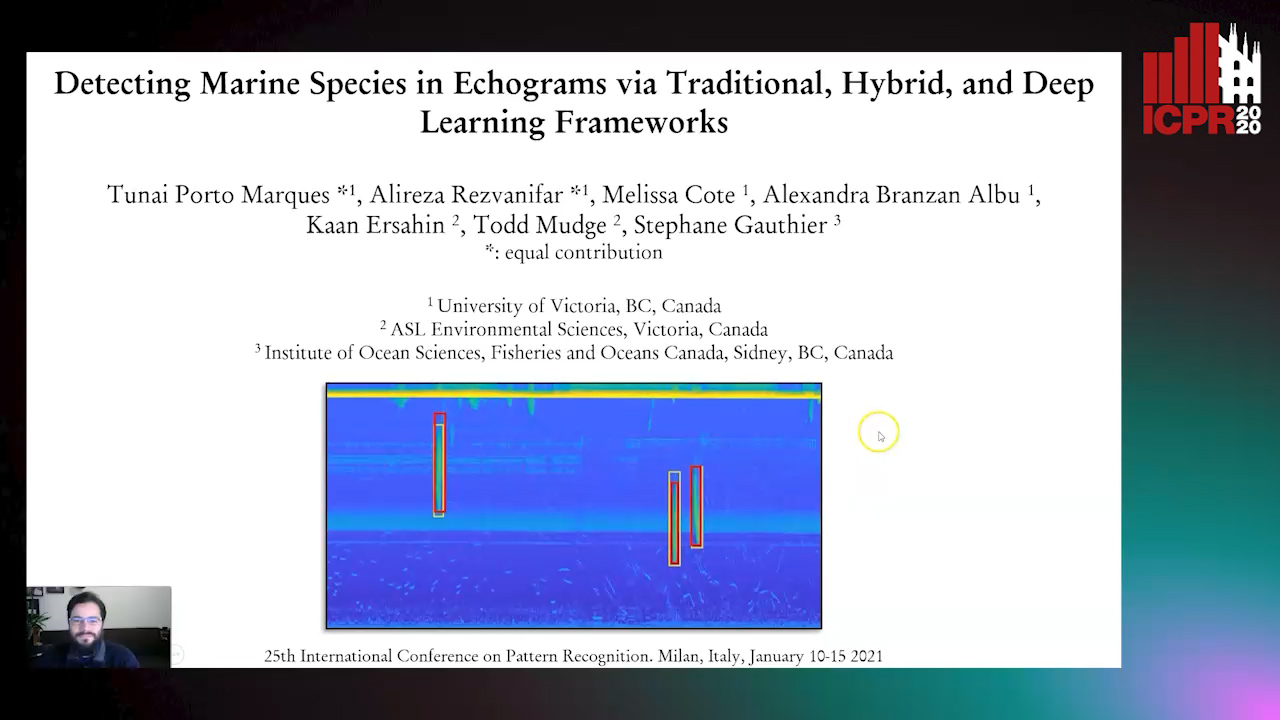
Auto-TLDR; End-to-End Deep Learning for Echogram Interpretation of Marine Species in Echograms
Abstract Slides Poster Similar
Do Not Treat Boundaries and Regions Differently: An Example on Heart Left Atrial Segmentation
Zhou Zhao, Elodie Puybareau, Nicolas Boutry, Thierry Geraud
Auto-TLDR; Attention Full Convolutional Network for Atrial Segmentation using ResNet-101 Architecture
Classify Breast Histopathology Images with Ductal Instance-Oriented Pipeline
Beibin Li, Ezgi Mercan, Sachin Mehta, Stevan Knezevich, Corey Arnold, Donald Weaver, Joann Elmore, Linda Shapiro

Auto-TLDR; DIOP: Ductal Instance-Oriented Pipeline for Diagnostic Classification
Abstract Slides Poster Similar
A Systematic Investigation on Deep Architectures for Automatic Skin Lesions Classification
Pierluigi Carcagni, Marco Leo, Andrea Cuna, Giuseppe Celeste, Cosimo Distante

Auto-TLDR; RegNet: Deep Investigation of Convolutional Neural Networks for Automatic Classification of Skin Lesions
Abstract Slides Poster Similar
Offset Curves Loss for Imbalanced Problem in Medical Segmentation
Ngan Le, Duc Toan Bui, Khoa Luu, Marios Savvides

Auto-TLDR; Offset Curves Loss for Medical Image Segmentation
On Morphological Hierarchies for Image Sequences
Caglayan Tuna, Alain Giros, François Merciol, Sébastien Lefèvre

Auto-TLDR; Comparison of Hierarchies for Image Sequences
Abstract Slides Poster Similar
CT-UNet: An Improved Neural Network Based on U-Net for Building Segmentation in Remote Sensing Images
Huanran Ye, Sheng Liu, Kun Jin, Haohao Cheng
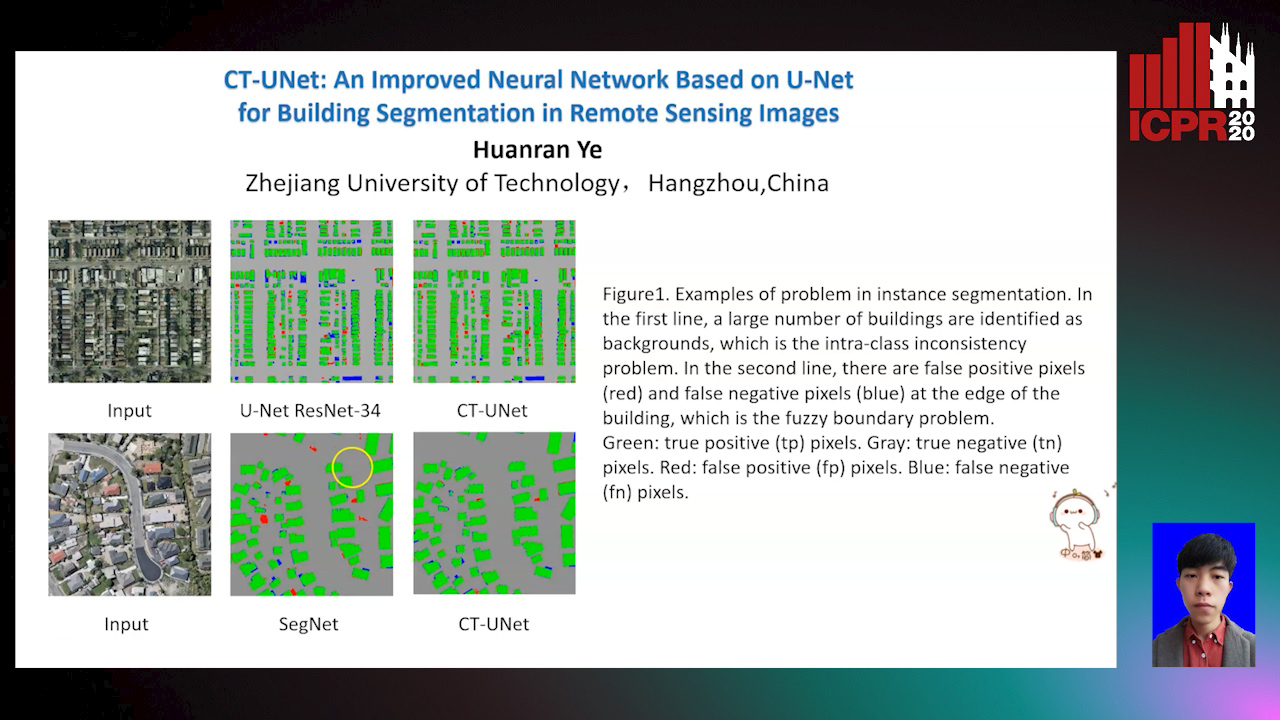
Auto-TLDR; Context-Transfer-UNet: A UNet-based Network for Building Segmentation in Remote Sensing Images
Abstract Slides Poster Similar
SAGE: Sequential Attribute Generator for Analyzing Glioblastomas Using Limited Dataset
Padmaja Jonnalagedda, Brent Weinberg, Jason Allen, Taejin Min, Shiv Bhanu, Bir Bhanu

Auto-TLDR; SAGE: Generative Adversarial Networks for Imaging Biomarker Detection and Prediction
Abstract Slides Poster Similar
Bridging the Gap between Natural and Medical Images through Deep Colorization
Lia Morra, Luca Piano, Fabrizio Lamberti, Tatiana Tommasi
Auto-TLDR; Transfer Learning for Diagnosis on X-ray Images Using Color Adaptation
Abstract Slides Poster Similar
BCAU-Net: A Novel Architecture with Binary Channel Attention Module for MRI Brain Segmentation
Yongpei Zhu, Zicong Zhou, Guojun Liao, Kehong Yuan

Auto-TLDR; BCAU-Net: Binary Channel Attention U-Net for MRI brain segmentation
Abstract Slides Poster Similar
Merged 1D-2D Deep Convolutional Neural Networks for Nerve Detection in Ultrasound Images
Mohammad Alkhatib, Adel Hafiane, Pierre Vieyres
Auto-TLDR; A Deep Neural Network for Deep Neural Networks to Detect Median Nerve in Ultrasound-Guided Regional Anesthesia
Abstract Slides Poster Similar
Fused 3-Stage Image Segmentation for Pleural Effusion Cell Clusters
Sike Ma, Meng Zhao, Hao Wang, Fan Shi, Xuguo Sun, Shengyong Chen, Hong-Ning Dai

Auto-TLDR; Coarse Segmentation of Stained and Stained Unstained Cell Clusters in pleural effusion using 3-stage segmentation method
Abstract Slides Poster Similar
Triplet-Path Dilated Network for Detection and Segmentation of General Pathological Images
Jiaqi Luo, Zhicheng Zhao, Fei Su, Limei Guo

Auto-TLDR; Triplet-path Network for One-Stage Object Detection and Segmentation in Pathological Images
Aerial Road Segmentation in the Presence of Topological Label Noise
Corentin Henry, Friedrich Fraundorfer, Eleonora Vig

Auto-TLDR; Improving Road Segmentation with Noise-Aware U-Nets for Fine-Grained Topology delineation
Abstract Slides Poster Similar
Deep Learning-Based Type Identification of Volumetric MRI Sequences
Jean Pablo De Mello, Thiago Paixão, Rodrigo Berriel, Mauricio Reyes, Alberto F. De Souza, Claudine Badue, Thiago Oliveira-Santos

Auto-TLDR; Deep Learning for Brain MRI Sequences Identification Using Convolutional Neural Network
Abstract Slides Poster Similar
NephCNN: A Deep-Learning Framework for Vessel Segmentation in Nephrectomy Laparoscopic Videos
Alessandro Casella, Sara Moccia, Chiara Carlini, Emanuele Frontoni, Elena De Momi, Leonardo Mattos

Auto-TLDR; Adversarial Fully Convolutional Neural Networks for kidney vessel segmentation from nephrectomy laparoscopic videos
Abstract Slides Poster Similar
One-Stage Multi-Task Detector for 3D Cardiac MR Imaging
Weizeng Lu, Xi Jia, Wei Chen, Nicolò Savioli, Antonio De Marvao, Linlin Shen, Declan O'Regan, Jinming Duan

Auto-TLDR; Multi-task Learning for Real-Time, simultaneous landmark location and bounding box detection in 3D space
Abstract Slides Poster Similar